Abstract
Toll-like receptors (TLR) are well-characterized pattern recognition receptors that can recognize and respond to diverse pathogen-associated or danger-associated molecular patterns during infection. TLR signaling in macrophages triggers in the intracellular signaling pathways through the recruitment of various adaptor and signaling proteins, and results in the activation of effector mechanisms and pathways that are important for host defense to intracellular bacteria. Effector mechanisms include inflammatory responses, cytokine generation, production of reactive oxygen species, and antimicrobial proteins. Accumulating studies showed that autophagy is a key pathway in the maintenance of homeostasis and housekeeping functions during infection and inflammation. In this review, we summarize the major effector pathways and mechanisms in the activation of TLR-inducible innate immune responses in macrophages. In addition, we focus the emerging evidence of crosstalk between autophagy and TLR-mediated signaling in terms of effector function of innate immune responses. A better understanding of effector functions by the activation of TLR-mediated signaling cascades contributes to the development of new therapeutics and vaccines against various intracellular pathogenic infections.
The innate immune response is a crucial component of the first-line defense system against a variety of invading pathogens (12). Macrophages are principal innate immune cell types that participate in recognition, signaling, digestion, antigen presentation, and effector functions in the infections to a variety of pathogens (23). Toll-like receptors (TLRs) are one of the best characterized innate immune receptors that can sense a variety of pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs) during bacterial and viral infections (145). In macrophages, TLR engagement by various PAMPs and DAMPs is importantly involved in the activation of intracellular signaling cascades that result in the activation of effector mechanisms and pathways to eradicate the invading pathogens (67). The activation of TLR responses triggers the sequential activation of signaling proteins and adaptors, effector enzymes through cooperative assembly mechanisms that elicit the innate immune responses (4578). The effector pathways of innate immune system consist of the expression of proinflammatory cytokines and chemokines, generation of reactive oxygen species (ROS) and nitrogen species, and production of antimicrobial proteins (79).
Autophagy is an intracellular catabolic process that can promote the destruction of invading pathogens in the autolysosomes (10). Accumulating evidence suggest that autophagy acts as a critical effector system in the host cells when they are invaded by pathogens (111213). Therefore, numerous bacteria and virus have evolved multiple strategies to exploit or evade from host defensive autophagy (101113). Indeed, TLR signaling can activate autophagy and lysosomal function, and LC3-associated phagocytosis (LAP) when combined with phagocytosis during infection, which contribute to innate effector function (1415). Since there are numerous studies for the role of autophagy in the regulation of inflammation (161718), it may contribute to control excessive inflammatory responses during pathogenic infection. We summarize the TLR-mediated activation of innate effector mechanisms and particularly focus on the role of autophagy during intracellular bacterial infection.
TLRs are the distinct pattern recognition receptor (PRR) members that include 10 and 12 family members in human and mice, respectively (8). TLR has a three-domain structure, leucine-rich repeats (LRRs) for the recognition of PAMPs, a transmembrane region, and an intracellular Toll/IL-1 receptor (TIR) domain for the signaling activation. In addition, TLRs are localized either in the plasma membrane (TLR1, TLR2, TLR4-6, and TLR10) or endosomal membranes (TLR3, TLR7-9, TLR11-13) (19). In addition, recent studies identified the molecules, such as uncoordinated 93 homolog B1 (UNC93B1) and leucine-rich repeat containing protein (LRRC) 59, for mediating endosomal TLR trafficking from ER to intracellular compartments (2021).
Upon the LRR engagement of TLRs by various PAMPs or DAMPs, TLRs trigger the recruitment of TIR domain-containing adaptors such as myeloid differentiation primary response protein 88 (MyD88) and TIR domain-containing adaptor protein inducing IFNβ (TRIF), TIR domain-containing adaptor protein (TIRAP)/MyD88 adapter-like protein (Mal), or TRIF-related adaptor molecule (TRAM). MyD88 is recruited by all TLR molecules, whereas TRIF is utilized by TLR3 and 4. The recruitment of MyD88 leads to the phosphorylation and activation of IL-1 receptor-associated kinase (IRAK) kinase family members including IRAK4 and IRAK1, which associates with E3 ubiquitin ligase TNF receptor associated factor (TRAF) 6 to activate the “master” TAK1 protein kinase complex (2223). Upon activation, TAK1 induces the activation of both NF-κB and mitogen-activated protein kinase (MAPK) pathways (ERK1/2, p38, and JNK) for further activation of transcriptional factors including AP-1 family members to produce proinflammatory cytokines including tumor necrosis factor (TNF)-α, interleukin (IL)-6, and various chemokines (824). In addition, the other adaptor TRIF interacts with TRAF6 and TRAF3, to facilitate the activation of TAK1 complex and TBK1 pathways, respectively. TRIF-dependent TRAF3 activation leads to the activation of type I interferon responses through phosphorylation of transcriptional factor IRF3 (82425).
During this sophisticated signaling activation, ubiquitination plays a key orchestrating role for controlling innate immune responses and inflammation through post-translational modification of target proteins (26). Both TRAF6 and TRAF3 are well-known and essential ubiquitin ligases that induce polyubiquitylation of target proteins to recruit for the activation or degradation during TLR signaling. Numerous studies identified crucial E3 ubiquitin ligases that play key regulatory roles in TLR signaling (27). For example, the Pellino family E3 ubiquitin ligases are involved in K63-linked polyubiquitylation of K63-linked polyubiquitination of IRAK1, TBK1, TAK1, receptor-interacting protein 1 (RIP1; also known as RIPK1) in TLR signaling pathways (272829). A recent study showed that the ubiquitin ligase RNF19A is importantly involved in the K48-linked ubiquitination and degradation of TRAF6, thereby attenuating TLR signaling (30). Figure 1 summarizes an overview of TLR-induced intracellular signaling pathways in innate immune cells. Deciphering the detailed mechanisms by which TLR signaling is regulated in macrophages may suggest future strategies to enhance host innate defense and prevent excessive inflammation.
Upon TLR activation, the intracellular signal transduction via numerous adaptors/enzymes leads to the activation of multiple effector mechanisms and pathways. Here we will review three major innate immune pathways, antimicrobial proteins, mitochondrial ROS, and auutophagy, in terms of TLR activation.
TLR-mediated innate immune signaling ultimately activates the exprssion of numerous antimicrobial peptides (AMPs) which are multifunctional molecules that kill pathogens, play immunomodulatory roles, and inflammation, in various settings of infection and inflammation (313233). Here we discuss two major types of AMPs, defensins and cathelicidins, in terms of TLR signaling.
Both defensins and cathelicidins are principal AMPs that can be synthesized in various cell types including innate immune cells, such as epithelial cells and macrophages. Previous studies suggest that TLR signaling leads to the differential profiling of AMP production depending on the specific cell or tissue types. Recent studies showed that the TLR4-mediated increase of β-defensin 2 expression in mesenchymal stem cells elicited antibacterial effects and down-regulated inflammatory responses during Escherichia coli-induced pneumonia (34). TLR stimulation contributed to the enhancement of host defense through the induction of nondefensin proteins, but not defensin-family proteins, to increase antibacterial activity in the intestinal mucosa after antibiotics treatment (35). In addition, there is a synergistic activation of antimicrobial peptide production when both TLRs and nucleotiden binding oligomerization domain 1 and 2 (NOD1 and NOD2) are stimulated together (36).
Recent studies provided the evidence for the opposite regulation of AMP in TLR-mediated immune responses that reshape the host defense and inflammation (31). For example, cathelicidins negatively regulate macrophage activation and inflammation through binding lipoproteins and LPS (37). LL-37, a C-terminal portion of human cathelicidin antimicrobial protein (hCAP18) (38), is known to decrease TLR4 signaling, but enhance TLR3 signaling (3940). Indeed, LL-37 can bind to various agonists of TLRs, and affect TLR-mediated signaling and bacterial phagocytosis (394041). In addition, there is a synergistic interaction between AMP and TLR signaling in the activation of cytokine production. Previous studies showed that human β-defensin-2 and -3 led to a synergistically increased production of inflammatory cytokines and chemokines in response to TLR ligands, through ATP release (42). Another study showed that human β-defensin 3 had an inhibitory function in the TLR4-mediated transcriptional activation of cathelicidon of proinflammatory genes in macrophages (43). Although it is not clear in terms of TLR signaling, the function of cathelicidins has been reported in the modulation of angiogenesis, presumably linked to tumorigenesis and inflammation (4445). The interaction of cathelicidins and defensins with microenvironment results in multifunctional roles in infection and inflammation (Fig. 2).
Importantly, TLR2/1 activation led to an induction of vitamin D signaling pathway and LL-37 production to enhance intracellular killing effects in human monocytes/macrophages against infection with Mycobacterium tuberculosis (Mtb), a major pathogen of human tuberculosis (TB) (46). In addition, TLR2/1 activation stimulated the induction of defensin-β4, which depends on IL-1β production in human monocytes (47). Moreover, TLR2-mediated hippo (mammalian sterile 20-like 1 and 2 kinases, MST1/2, inmammals) signaling is required for the paracrine activation of antimicrobial peptide β-defensins through CXCL1 and CXCL2 secretion during Mtb infection (48). These data suggest that TLR signaling cooperates with other signaling pathways for the induction of AMP to contribute to TLR-mediated antimicrobial host defense.
Recent studies provided evidence that mitochondria are crucial organelles to regulate innate immune responses to various DAMP and PAMP signals (49). In macrophages, activation of TLR1, 2, and 4 are associated with the increased production of mitochondrial ROS, which are critically involved in bactericidal activities and the recruitment of mitochondria to bacterial phagosomes (50). This response is mediated by the translocation of the TRAF6 to the mitochondria, and the TRAF6 interaction with ECSIT (evolutionary conserved signaling intermediate in Toll pathways), a protein that is esential for the respiratory chain assembly and increased mitochondrial ROS generation (50). A more recent paper revealed the mechanisms for mitochondrial trafficking and juxtaposition to bacterial phagosomes are mediated by the kinases Mst1 and Mst2 via the activation of the Rho family GTPase Rac (51). The GTPase Rac activation is required for the TLR-mediated interaction of TRAF6-ECSIT, thereby recruiting mitochondria to phagosomes and subsequent killing of microbes (51). Interestingly, peroxiredoxin-6, an antioxidant enzyme of peroxiredoxin family, is required for the inhibition of mitochondrial ROS production through interruption of the TRAF6-ECSIT complex formation in response to TLR4 (52). In addition, a recent paper showed another function of mitochondrial ROS production that is required for the antigen cross-presentation capacity of plasmacytoid dendritic cells after TLR stimulation (53).
Autophagy is a lysosomal degradation pathway and acts as an innate immune effector because it mediates the clearance of intracellular microbes (5455). In addition, autophagy plays an important role in various aspects of immune responses, including antigen presentation, regulation of cytokine production, and lymphocyte homeostasis (55). Here we briefly discuss the overview of autophagy, TLR-induced autophagy and its consequences during infection, and the mechanisms for crosstalk between TLR signaling and autophagy process.
Autophagy process is required for intracellular maintenance of homeostasis during various stress signals including nutrient starvation and pathogenic invasion (5657). The autophagic process is divided at least three steps: initiation, elongation, and maturation. In the initiation step, cargos are surrounded by a cup-shaped double-membrane structure, phagophore, through the action of ULK1/2 complex (ULK1/2-Atg13-Atg101-FIP200) and Class III PI3K-Beclin-1 complex (Beclin 1-ATG14L-Vps34-p150). The phagophore structure expands to autophagosomes, through two ubiquitin-like conjugation systems containing core autophagy proteins including Atg8/LC3 and Atg5-Atg12-Atg16L1 (56). In the autophagosome maturation step, the autophagosomal structures can be fused with endolysosomal vesicles to form degradative autolysosomes through several proteins including SNAREs, ATG8 family members, and Rab GTPases (58). Since autophagy is essential in the maintenance of house-keeping function and homeostasis, its dysfunction and altered regulation are associated with numerous diseases (59).
Although autophagy was considered to be a nonspecific response, it is now being clear that autophagy can target specific organelles or substances including pathogens, i.e., xenophagy (57). Numerous autophagic adaptors including p62, NDP52, and optineurin, play an important role in the activation of selective autophagy activation (606162). Since the autophagic adaptors have both domains, i.e., LC3-interacting region and ubiquitin-binding domains, to connect ubiquitinylated cargos to autophagic machinery (6061). Numerous pathogens have evolved different mechanisms to escape or exploit autophagy/xenophagy to get an advantage for the survival of pathogens (63). Here we briefly introduce xenophagy against Mtb infection, because xenophagy has been widely studied in Mtb infection.
Mtb can reside in the phagosomes in macrophages, but access into the cytosols through ESX-1 system (64). Mtb and their DNA can be recognized by cytosolic sensor c-GAS, and subsequently ubiquinylated by Parkin and Smurf1 (656667). Xenophagy against Mtb is mediated through autophagic adaptors including p62 and NDP52 (64). In addition, TRIM and Galectin family proteins cooperate the recognition of damaged phagosomes, thereby the core autophagy proteins can be targeted to activate xenophagy process (6869). In this process, the mycobactericidal activity is presumably due to the formation of neo-antimicrobial peptides, which are delivered from numerous bulk ubiquitinated proteins during autophagic process, through the action of p62 (7071). In human monocytes/macrophages, vitamin D3-induced autophagy activation was essentially required for antimicrobial responses through induction of cathelicidins (727374). In addition, IRGM, a human immunity-related GTPase, induces xenophagy through interaction with ULK1 and Beclin 1, to enhance the formation of autophagy initiation complex and antimicrobial functions (75). Figure 3 summarizes a brief overview of autophagy and xenophagy during Mtb infection.
Although there are a large body of evidence that autophagy/xenophagy promotes antimicrobial responses to Mtb infection, recent studies argued about the function of autophagy in vivo. Genetic deletion models of autophagy genes in myeloid lineage of the mice did not exhibit protective effects against Mtb infection in vivo, except Atg5 that showed an autophagy-independent, and controlling neutrophil-mediated harmful inflammation to the host (76). Despite this, autophagy may contribute to the protective immune responses and improvement of antigen-presentation, thus being a promising target for the development of new vaccines and therapeutics (77).
TLR signaling activation in innate immune cells results in various effector mechanisms/pathways including autophagy process, during pathogenic infection (78). Previous studies showed that TLR signaling activation led to the induction of autophagy in macrophages (7980). Among TLRs, TLR7 activation showed strong effects upon the enhancement of autophagy leading to the elimination of intracelular microbes in macrophages (79). TLR7-mediated autophagy depended on MyD88 expression (79). In addition, TLR4-induced autophagy was mediated through TRIF-dependent, but MyD88-independent, pathway (81). Interestingly, both TLR4- and TLR7-mediated autophagy activation resulted in the increased colocalization of mycobacterial phagosomes and autophagosomes, indicating a promotion of phagosomal maturation (8081).
In human monocytes/macrophages, TLR2/1 activation activated AMPK-dependent functional vitamin D signaling activation, which leads to the induction of autophagy activation and antimicrobial responses (82). Another study showed that treatment of murine macrophages with TLR4/LPS or TLR3/poly(I:C) resulted in the elimination of intracellular mycobacteria through autophagy activation (83). Either stimulation of TLR2 or TLR4 enhanced the expression of a serine protease inhibitor plasminogen activator inhibitor type 2 (PAI-2), which stabilized Beclin 1 to activate autophagy pathway to suppress NLRP3 activation (84). Thus TLR-mediated autophagy activation may contribute to cell-autonomous antimicrobial defense and controlling excessive inflammation. In other aspect, TLR signaling plays a negative regulatory role in autophagy activation in different context. Recent studies showed that TLR4/LPS stimulation inhibited autophagy in microglial cells through negative regulation of FOXO3 at downstream of PI3K pathway (85). In Leishmania infection, TLR3, 7, 9, as well as UNC93B1, a chaperone for trafficking of nucleic-sensing TLRs to endolysosomes, are importantly required for the activation of anti-microbial autophagy and controlling parasite replication (86).
TLR8 stimulation robustly activated the expression of genes involved in vitamin D signaling, which promoted vitamin D-cathelicidin-dependent autophagy and antimicrobial responses against human immunodeficiency virus infection in human macrophages (87). Thus TLR signaling may adopt distinct mechanisms and effects upon autophagy in the different context of manner. A brief overview that TLR signaling regulates antibacterial autophagy is shown in Figure 4. In addition, TLR4 stimulation led to an induction of LC3-positive structures, which are independent of canonical macroautophagy and contain p62 as a component (88). The formation of p62-positive selective autophagosome was mediated through Nrf2 and TLR4-MyD88 signaling pathways (88). These data suggest that TLR-mediated innate immune and ROS pathways converge to autophagy activation that contribute to promote host defense during infection.
There are several evidence that the autophagy pathway regulates TLR-mediated innate immune responses, suggesting there is a complex link between TLR responses and autophagy. For instance the formation of immunoamphisome in plasmacytoid dendritic cells enhanced TLR7-mediated responses and antigen presentation on both MHC class I and class II molecules (89). In intestinal epithelial cells, TLR-induced autophagy and Atg7 expression are required for IL-8 production, suggesting a role for autophagy in intestinal innate immune responses (90). In contrast, suppression of autophagy pathway by either pharmacological inhibitors of autophagy or RNA interference amplified the IL-23 production in macrophages and dendritic cells in response to TLR activation (91).
A large body of evidence showed that there is intimate connection between TLR signaling-related molecules and autophagy-related proteins. Earlier studies showed that TLR family adaptors MyD88 and TRIF were associated with Beclin-1, a key autophagy-related protein in the autophagosome formation (92). It was also reported that TRAF6 was essentially involved in the K63-linked polyubiquitination of Beclin-1 to induce TLR4-mediated autophagy in macrophages (9394). In addition, a well-known deubiqutinase A20 suppressed the K63-linked polyubiquitination of Beclin-1 to inhibit the autophagosome formation (9394). During Pseudomonas aeruginosa infection, NLRC4 inflammasome-dependent caspase-1 activation led to a cleavage of TRIF, thus attenuating autophagy activation and type I interferon expression (95). In vivo infection model with P. aeruginosa showed that TRIF was required for the enhancement of antibacterial autophagy and clearance of bacteria (95). A recent study showed that the TLR adaptor molecule Mal (encoded by TIRAP) through IFN-γ receptor signaling, which in turn activated autophagy and increased the protection from M. tuberculosis infection (96). This study is important for the potential explanation of genetic variant of Mal (S180L polymorphism; murine equivalent S200L) in the increased susceptibility to TB (96). Another recent study revealed that TRIF degradation, which was mediated through selective autophagic pathway via the E3 ligase TRIM32 and the autophagic adaptor TAX1BP1, negatively regulated TLR3/4-induced type I interferon and proinflammatory immune responses (97).
There are several reports that autophagy proteins are involved in innate immune responses associated with TLR signaling. In mycobacterial infection models, the autophagy protein DNA damage-regulated autophagy modulator (DRAM) 1 expression was dependent on MyD88- and NF-κB-mediated innate immune signaling in human macrophages and zebrafish embryos (98). Moreover, the autophagy inhibitor Rubicon played a role as a feedback regulator of CARD9, BCL10, and MALT1 (CBM complex)-mediated innate immune signaling (99). Rubicon functioned in the disassembly of CBM complex formation and inhibited cytokine production (99). In addition, TLR2 activation led to an interaction of Rubicon with the p22phox of NADPH-oxidase complex for the enhancement of antimicrobial effects through induction of ROS and inflammatory cytokine generation (100). A recent study showed that Rubicon plays an essential role for antiviral type I interferon responses through interaction with interferon regulatory factor (IRF) 3 and inhibition of IRF3 dimerization (101). Together, these studies indicate an essential role for TLR signaling adaptors in the regulation of the autophagy protein function or vice versa.
Considerable progress has been made in elucidating the mechanisms for TLR-induced signaling in innate immunity. For innate immune cells such as macrophages, the activation of TLR-mediated innate immune signaling leads to the effector pathways including cytokine generation, reactive nitrogen and oxygen species, production of antimicrobial proteins, autophagy, etc. Although a great advance has been done in the unveiling the signaling mechanisms by which TLR activation leads to the innate immune responses, many questions remained in the functional identification of positive and negative regulators in TLR-mediated signaling pathways.
In addition to providing a signaling map of TLR pathway, future studies are warranted to investigate the immunomodulatory functions of diverse antimicrobial proteins in innate immune responses. Numerous efforts support a role for mitochondrial ROS in the antimicrobial responses against pathogens. However, dysregulation of ROS generation could amplify chronic inflammation during infection. Thus it remains to be seen whether TLR-mediated autophagy or other cellular pathway is required for the coordinated regulation of innate effector responses.
The information on the function of autophagy/xenophagy is emerging in a variety of infection; one example would be Mtb-induced host defensive responses. Accumulating evidence suggests that TLR signaling activates autophagy, which affects TLR-mediated innate immune responses during infection. Understanding the molecular mechanisms for biological connections between autophagy and TLR signaling is a challenge for the possibility of novel therapeutics against infection and inflammation.
Figures and Tables
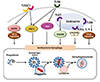 | Figure 1An overview of TLR-induced intracellular signaling pathways in innate immune cells. Upon TLR signaling, the intracellular signaling cascades involve the actions of numerous adaptors MyD88, TRIF, MAL/TIRARAP, or TRAM, and multiple signaling molecules. In particular, MyD88 is recruited by all TLR molecules, whereas TRIF is utilized by TLR3 and 4. The recruitment of MyD88 activates IRAK kinase family members, which associates with E3 ubiquitin ligase TRAF6 to activate TAK1 protein kinase complex. This leads to the activation of both NF-κB and mitogen-activated protein kinase (MAPK) pathways (ERK1/2, p38, and JNK) for proinflammatory cytokine generation. The other adaptor TRIF interacts with both TRAF6 and TRAF3, to facilitate the activation of TAK1 complex and TBK1 pathways, leading to the activation of type I interferon responses. In the sophisticated process, uncoordinated 93 homolog B1 (UNC93B 1) and leucine-rich repeat containing protein (LRRC) 59, are required for the endosomal TLR trafficking from ER to endosomal structures. |
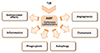 | Figure 2Multiple functions of antimicrobial proteins in innate immune responses. TLR-induced intracellular signaling pathways culminate into the activation of innate effector pathways including the generation of antimicrobial proteins. Two important antimicrobial peptides, cathelicidins and defensins, function in various aspects of biological responses, i.e., the regulation of inflammation, phagocytosis, autophagy, as well as antimicrobial effects, in innate immune cells. |
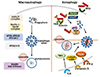 | Figure 3A schematic overview of macroautophagy and xenophagy in mycobacterial infection. Macroautophagy process include three steps: initiation (formation of phagophore), elongation into double-membraned autophagosomal structure, and its maturation by lysosomal fusion. Xenophagy process is well-characterized in mycobacterial infection. During Mtb infection, xenophagy is activated by cytoplasmic release of Mtb, which can be ubiquitinated by E3 ligases Parkin and Smurf1. Then ubiquitinated Mtb phagosomes are recognized by autophagic receptors p62 and NDP52, which contain domains for interaction with ubiquitinated cargos and LC3-containing autophagic machinery. It is also well-known that, several innate immune signals including TLR, vitamin D receptor signaling, IFN-γh distinct mechanisms involving IRGM (for IFN-γ), cathelicidins (for vitamin D receptor signaling), and TBK1 (for TLR) in monocytes/macrophages. |
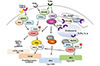 | Figure 4Figure 4. TLR-mediated regulation of antibacterial autophagy. TLR signaling activation in innate immune cells results in the activation of antibacterial autophagy. TLR7 signaling activates autophagy through MyD88; TLR4 stimulation induces autophagy through a TRIF-dependent pathway. TLR2/1 stimulation activates AMPK-dependent functional vitamin D signaling activation, leading to the induction of autophagy activation and antimicrobial responses. Either TLR2 or TLR4 stimulation activates autophagy through a serine protease inhibitor PAI-2 via stabilization of Beclin 1. In addition, TLR3, 7, 9, and UNC93B1, are required for the activation of antibacterial autophagy. TLR8 stimulation activates the expression of genes involved in vitamin D signaling and cathelicidin-dependent autophagy in human macrophages. |
ACKNOWLEDGEMENTS
We thank Dr. Silwal P for critical reading of paper. We are indebted to current and past members of our laboratory for discussions and investigations that contributed to this article. This work was supported by the Chungnam National University. I apologize to colleagues whose work and publications could not be referenced owing to space constraints. The authors have no financial conflict of interests.
References
1. Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001; 2:675.


2. Hu X, Chakravarty SD, Ivashkiv LB. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol Rev. 2008; 226:41–56.



3. Wynn TA, Barron L. Macrophages: master regulators of inflammation and fibrosis. Semin Liver Dis. 2010; 30:245–257.



4. Cui J, Chen Y, Wang HY, Wang RF. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum Vaccin Immunother. 2014; 10:3270–3285.


6. Gulati A, Kaur D, Krishna Prasad GVR, Mukhopadhaya A. PRR function of innate immune receptors in recognition of bacteria or bacterial ligands. Adv Exp Med Biol. 2018; 1112:255–280.


7. Vajjhala PR, Ve T, Bentham A, Stacey KJ, Kobe B. The molecular mechanisms of signaling by cooperative assembly formation in innate immunity pathways. Mol Immunol. 2017; 86:23–37.


9. Nanson JD, Kobe B, Ve T. Death, TIR, and RHIM: Self-assembling domains involved in innate immunity and cell-death signaling. J Leukoc Biol. 2019; 105:363–375.


10. Campoy E, Colombo MI. Autophagy in intracellular bacterial infection. Biochim Biophys Acta. 2009; 1793:1465–1477.


11. Borel S, Espert L, Biard-Piechaczyk M. Macroautophagy regulation during HIV-1 infection of CD4+ T cells and macrophages. Front Immunol. 2012; 3:97.



12. Deretic V. Autophagy, an immunologic magic bullet: Mycobacterium tuberculosis phagosome maturation block and how to bypass it. Future Microbiol. 2008; 3:517–524.



13. Bradfute SB, Castillo EF, Arko-Mensah J, Chauhan S, Jiang S, Mandell M, et al. Autophagy as an immune effector against tuberculosis. Curr Opin Microbiol. 2013; 16:355–365.



14. Basu J, Shin DM, Jo EK. Mycobacterial signaling through toll-like receptors. Front Cell Infect Microbiol. 2012; 2:145.



15. Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007; 450:1253–1257.


16. Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018; 14:243–251.



17. Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013; 13:722–737.



18. Fésüs L, Demény MÁ, Petrovski G. Autophagy shapes inflammation. Antioxid Redox Signal. 2011; 14:2233–2243.


19. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010; 11:373–384.


20. Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, Schekman R, et al. UNC93B1 mediates differential trafficking of endosomal TLRs. Elife. 2013; 2:e00291.

21. Tatematsu M, Funami K, Ishii N, Seya T, Obuse C, Matsumoto M. LRRC59 regulates trafficking of nucleic acid-sensing TLRs from the endoplasmic reticulum via association with UNC93B1. J Immunol. 2015; 195:4933–4942.


23. Ajibade AA, Wang HY, Wang RF. Cell type-specific function of TAK1 in innate immune signaling. Trends Immunol. 2013; 34:307–316.


24. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006; 124:783–801.


25. Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010; 11:70–75.


26. Zinngrebe J, Montinaro A, Peltzer N, Walczak H. Ubiquitin in the immune system. EMBO Rep. 2014; 15:28–45.


27. Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol. 2011; 12:35–48.


28. Murphy M, Xiong Y, Pattabiraman G, Qiu F, Medvedev AE. Pellino-1 positively regulates toll-like receptor (TLR) 2 and TLR4 signaling and is suppressed upon induction of endotoxin tolerance. J Biol Chem. 2015; 290:19218–19232.



29. Medvedev AE, Murphy M, Zhou H, Li X. E3 ubiquitin ligases Pellinos as regulators of pattern recognition receptor signaling and immune responses. Immunol Rev. 2015; 266:109–122.



30. Wu C, Su Z, Lin M, Ou J, Zhao W, Cui J, et al. NLRP11 attenuates toll-like receptor signalling by targeting TRAF6 for degradation via the ubiquitin ligase RNF19A. Nat Commun. 2017; 8:1977.

31. Lee EY, Lee MW, Wong GCL. Modulation of toll-like receptor signaling by antimicrobial peptides. Semin Cell Dev Biol. 2018; 88:173–184.



33. Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol. 2012; 32:143–171.


34. Sung DK, Chang YS, Sung SI, Yoo HS, Ahn SY, Park WS. Antibacterial effect of mesenchymal stem cells against Escherichia coli is mediated by secretion of beta-defensin-2 via toll-like receptor 4 signalling. Cell Microbiol. 2016; 18:424–436.


35. Wu YY, Hsu CM, Chen PH, Fung CP, Chen LW. Toll-like receptor stimulation induces nondefensin protein expression and reverses antibiotic-induced gut defense impairment. Infect Immun. 2014; 82:1994–2005.



36. Pashenkov MV, Murugina NE, Budikhina AS, Pinegin BV. Synergistic interactions between NOD receptors and TLRs: Mechanisms and clinical implications. J Leukoc Biol. 2019; 105:669–680.


37. Coorens M, Schneider VAF, de Groot AM, van Dijk A, Meijerink M, Wells JM, et al. Cathelicidins inhibit Escherichia coli-induced TLR2 and TLR4 activation in a viability-dependent manner. J Immunol. 2017; 199:1418–1428.



38. Sørensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, et al. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001; 97:3951–3959.


39. Lai Y, Adhikarakunnathu S, Bhardwaj K, Ranjith-Kumar CT, Wen Y, Jordan JL, et al. LL37 and cationic peptides enhance TLR3 signaling by viral double-stranded RNAs. PLoS One. 2011; 6:e26632.

40. Lai Y, Yi G, Chen A, Bhardwaj K, Tragesser BJ, Rodrigo AV, et al. Viral double-strand RNA-binding proteins can enhance innate immune signaling by toll-like Receptor 3. PLoS One. 2011; 6:e25837.

41. Wan M, van der Does AM, Tang X, Lindbom L, Agerberth B, Haeggström JZ. Antimicrobial peptide LL-37 promotes bacterial phagocytosis by human macrophages. J Leukoc Biol. 2014; 95:971–981.


42. Wanke D, Mauch-Mucke K, Holler E, Hehlgans T. Human beta-defensin-2 and -3 enhance pro-inflammatory cytokine expression induced by TLR ligands via ATP-release in a P2X7R dependent manner. Immunobiology. 2016; 221:1259–1265.


43. Semple F, MacPherson H, Webb S, Cox SL, Mallin LJ, Tyrrell C, et al. Human beta-defensin 3 affects the activity of pro-inflammatory pathways associated with MyD88 and TRIF. Eur J Immunol. 2011; 41:3291–3300.



44. Khurshid Z, Naseem M, Yahya IAF, Mali M, Sannam Khan R, Sahibzada HA, et al. Significance and diagnostic role of antimicrobial cathelicidins (LL-37) peptides in oral health. Biomolecules. 2017; 7:pii: E80.

45. Patel S, Akhtar N. Antimicrobial peptides (AMPs): The quintessential ‘offense and defense’ molecules are more than antimicrobials. Biomed Pharmacother. 2017; 95:1276–1283.


46. Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006; 311:1770–1773.


47. Liu PT, Schenk M, Walker VP, Dempsey PW, Kanchanapoomi M, Wheelwright M, et al. Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses. PLoS One. 2009; 4:e5810.
48. Boro M, Singh V, Balaji KN. Mycobacterium tuberculosis-triggered Hippo pathway orchestrates CXCL1/2 expression to modulate host immune responses. Sci Rep. 2016; 6:37695.



50. West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011; 472:476–480.



51. Geng J, Sun X, Wang P, Zhang S, Wang X, Wu H, et al. Kinases Mst1 and Mst2 positively regulate phagocytic induction of reactive oxygen species and bactericidal activity. Nat Immunol. 2015; 16:1142–1152.



52. Min Y, Wi SM, Shin D, Chun E, Lee KY. Peroxiredoxin-6 negatively regulates bactericidal activity and NF-kappaB activity by interrupting TRAF6-ECSIT complex. Front Cell Infect Microbiol. 2017; 7:94.



53. Oberkampf M, Guillerey C, Mouriès J, Rosenbaum P, Fayolle C, Bobard A, et al. Mitochondrial reactive oxygen species regulate the induction of CD8(+) T cells by plasmacytoid dendritic cells. Nat Commun. 2018; 9:2241.



54. Shibutani ST, Saitoh T, Nowag H, Münz C, Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol. 2015; 16:1014–1024.


55. Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012; 30:611–646.


56. Siqueira MDS, Ribeiro RM, Travassos LH. Autophagy and its interaction with intracellular bacterial pathogens. Front Immunol. 2018; 9:935.



58. Zhao YG, Zhang H. Autophagosome maturation: An epic journey from the ER to lysosomes. J Cell Biol. 2019; 218:757–770.



59. Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell. 2016; 3:588–596.



60. Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015; 282:4672–4678.


62. Viret C, Rozières A, Faure M. Novel insights into NDP52 autophagy receptor functioning. Trends Cell Biol. 2018; 28:255–257.


63. Castrejón-Jimenèz NS, Leyva-Paredes K, Hernández-González JC, Luna-Herrera J, García-Pèrez BE. The role of autophagy in bacterial infections. Biosci Trends. 2015; 9:149–159.


64. Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012; 150:803–815.



65. Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013; 501:512–516.



66. Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe. 2015; 17:811–819.



67. Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, et al. The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe. 2017; 21:59–72.


68. Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, et al. TRIMs and Galectins globally cooperate and TRIM16 and Galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev Cell. 2016; 39:13–27.



69. Kumar S, Chauhan S, Jain A, Ponpuak M, Choi SW, Mudd M, et al. Galectins and TRIMs directly interact and orchestrate autophagic response to endomembrane damage. Autophagy. 2017; 13:1086–1087.



70. Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010; 32:329–341.



71. Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007; 104:6031–6036.



72. Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, et al. Vitamin D3 induces autophagy in human monocytes/macrophages via cathelicidin. Cell Host Microbe. 2009; 6:231–243.


73. Rekha RS, Rao Muvva SS, Wan M, Raqib R, Bergman P, Brighenti S, et al. Phenylbutyrate induces LL-37-dependent autophagy and intracellular killing of Mycobacterium tuberculosis in human macrophages. Autophagy. 2015; 11:1688–1699.



74. Høyer-Hansen M, Nordbrandt SP, Jäättelä M. Autophagy as a basis for the health-promoting effects of vitamin D. Trends Mol Med. 2010; 16:295–302.


75. Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015; 58:507–521.



76. Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015; 528:565–569.



77. Paik S, Kim JK, Chung C, Jo EK. Autophagy: a new strategy for host-directed therapy of tuberculosis. Virulence. 2018; 15:1–12.

78. Stocks CJ, Schembri MA, Sweet MJ, Kapetanovic R. For when bacterial infections persist: Toll-like receptor-inducible direct antimicrobial pathways in macrophages. J Leukoc Biol. 2018; 103:35–51.


79. Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008; 27:1110–1121.



80. Delgado MA, Deretic V. Toll-like receptors in control of immunological autophagy. Cell Death Differ. 2009; 16:976–983.



81. Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007; 27:135–144.



82. Shin DM, Yuk JM, Lee HM, Lee SH, Son JW, Harding CV, et al. Mycobacterial lipoprotein activates autophagy via TLR2/1/CD14 and a functional vitamin D receptor signalling. Cell Microbiol. 2010; 12:1648–1665.



83. Xu Y, Fattah EA, Liu XD, Jagannath C, Eissa NT. Harnessing of TLR-mediated autophagy to combat mycobacteria in macrophages. Tuberculosis (Edinb). 2013; 93:S33–S37.

84. Chuang SY, Yang CH, Chou CC, Chiang YP, Chuang TH, Hsu LC. TLR-induced PAI-2 expression suppresses IL-1beta processing via increasing autophagy and NLRP3 degradation. Proc Natl Acad Sci U S A. 2013; 110:16079–16084.



85. Lee JW, Nam H, Kim LE, Jeon Y, Min H, Ha S, et al. TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy. 2018; DOI: 10.1080/15548627.2018.1556946. [Epub ahead of print].

86. Franco LH, Fleuri AKA, Pellison NC, Quirino GFS, Horta CV, de Carvalho RVH, et al. Autophagy downstream of endosomal Toll-like receptor signaling in macrophages is a key mechanism for resistance to Leishmania major infection. J Biol Chem. 2017; 292:13087–13096.



87. Campbell GR, Spector SA. Toll-like receptor 8 ligands activate a vitamin D mediated autophagic response that inhibits human immunodeficiency virus type 1. PLoS Pathog. 2012; 8:e1003017.

88. Fujita K, Maeda D, Xiao Q, Srinivasula SM. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci U S A. 2011; 108:1427–1432.



89. Blanchet FP, Piguet V. Immunoamphisomes in dendritic cells amplify TLR signaling and enhance exogenous antigen presentation on MHC-II. Autophagy. 2010; 6:816–818.


90. Li YY, Ishihara S, Aziz MM, Oka A, Kusunoki R, Tada Y, et al. Autophagy is required for toll-like receptor-mediated interleukin-8 production in intestinal epithelial cells. Int J Mol Med. 2011; 27:337–344.


91. Peral de, Jones SA, Ni Cheallaigh C, Hearnden CA, Williams L, Winter J, et al. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol. 2012; 189:4144–4153.


92. Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008; 283:33175–33182.



93. Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010; 3:ra42.

94. Shi CS, Kehrl JH. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy. 2010; 6:986–987.



95. Jabir MS, Ritchie ND, Li D, Bayes HK, Tourlomousis P, Puleston D, et al. Caspase-1 cleavage of the TLR adaptor TRIF inhibits autophagy and beta-interferon production during Pseudomonas aeruginosa infection. Cell Host Microbe. 2014; 15:214–227.


96. Ní Cheallaigh C, Sheedy FJ, Harris J, Muñnoz-Wolf N, Lee J, West K, et al. A Common Variant in the Adaptor Mal Regulates Interferon Gamma Signaling. Immunity. 2016; 44:368–379.



97. Yang Q, Liu TT, Lin H, Zhang M, Wei J, Luo WW, et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017; 13:e1006600.

98. van der Vaart M, Korbee CJ, Lamers GE, Tengeler AC, Hosseini R, Haks MC, et al. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense. Cell Host Microbe. 2014; 15:753–767.


99. Yang CS, Rodgers M, Min CK, Lee JS, Kingeter L, Lee JY, et al. The autophagy regulator Rubicon is a feedback inhibitor of CARD9-mediated host innate immunity. Cell Host Microbe. 2012; 11:277–289.







 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download