1. Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, et al. The global burden of cancer 2013. JAMA Oncol. 2015; 1:505–527. PMID:
26181261.
2. Park JY, Jang SH. Epidemiology of lung cancer in Korea: recent trends. Tuberc Respir Dis. 2016; 79:58–69.

3. Shin A, Oh CM, Kim BW, Woo H, Won YJ, Lee JS. Lung cancer epidemiology in Korea. Cancer Res Treat. 2017; 49:616–626. PMID:
27669705.

4. Balgkouranidou I, Liloglou T, Lianidou ES. Lung cancer epigenetics: emerging biomarkers. Biomark Med. 2013; 7:49–58. PMID:
23387484.

5. Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015; 16:e342–e351. PMID:
26149886.

6. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011; 12:861–874. PMID:
22094949.

7. Zhang Y. Recent progress in the epigenetics and chromatin field. Cell Res. 2011; 21:373–374. PMID:
21383679.

8. Ansari J, Shackelford RE, El-Osta H. Epigenetics in non-small cell lung cancer: from basics to therapeutics. Transl Lung Cancer Res. 2016; 5:155–171. PMID:
27186511.

9. Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in lung cancer diagnosis and therapy. Cancer Metastasis Rev. 2015; 34:229–241. PMID:
25939322.

10. Hong Y, Ji W, An S, Han SS, Lee SJ, Kim WJ. Sex differences of COPD phenotypes in nonsmoking patients. Int J Chron Obstruct Pulmon Dis. 2016; 11:1657–1662. PMID:
27524891.

11. Oken MM, Hocking WG, Kvale PA, Andriole GL, Buys SS, Church TR, et al. Screening by chest radiograph and lung cancer mortality: the Prostate, Lung, Colorectal, and Ovarian (PLCO) randomized trial. JAMA. 2011; 306:1865–1873. PMID:
22031728.
12. National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011; 365:395–409. PMID:
21714641.

13. van der Aalst CM, Ten Haaf K, de Koning HJ. Lung cancer screening: latest developments and unanswered questions. Lancet Respir Med. 2016; 4:749–761. PMID:
27599248.

14. Hubers AJ, Brinkman P, Boksem RJ, Rhodius RJ, Witte BI, Zwinderman AH, et al. Combined sputum hypermethylation and eNose analysis for lung cancer diagnosis. J Clin Pathol. 2014; 67:707–711. PMID:
24915850.

15. Kneip C, Schmidt B, Seegebarth A, Weickmann S, Fleischhacker M, Liebenberg V, et al.
SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011; 6:1632–1638. PMID:
21694641.
16. Ahrendt SA, Chow JT, Xu LH, Yang SC, Eisenberger CF, Esteller M, et al. Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J Natl Cancer Inst. 1999; 91:332–339. PMID:
10050866.

17. Xiao P, Chen JR, Zhou F, Lu CX, Yang Q, Tao GH, et al. Methylation of P16 in exhaled breath condensate for diagnosis of non-small cell lung cancer. Lung Cancer. 2014; 83:56–60. PMID:
24268095.

18. Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008; 358:1118–1128. PMID:
18337602.

19. Sandoval J, Mendez-Gonzalez J, Nadal E, Chen G, Carmona FJ, Sayols S, et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol. 2013; 31:4140–4147. PMID:
24081945.

20. Hsu HS, Chen TP, Hung CH, Wen CK, Lin RK, Lee HC, et al. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer. 2007; 110:2019–2026. PMID:
17876837.

21. Belinsky SA, Grimes MJ, Casas E, Stidley CA, Franklin WA, Bocklage TJ, et al. Predicting gene promoter methylation in non-small-cell lung cancer by evaluating sputum and serum. Br J Cancer. 2007; 96:1278–1283. PMID:
17406356.

22. Stirzaker C, Taberlay PC, Statham AL, Clark SJ. Mining cancer methylomes: prospects and challenges. Trends Genet. 2014; 30:75–84. PMID:
24368016.

23. Bibikova M, Le J, Barnes B, Saedinia-Melnyk S, Zhou L, Shen R, et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009; 1:177–200. PMID:
22122642.
24. Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. Critical evaluation of the Illumina Methylation-EPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016; 17:208. PMID:
27717381.

25. Qiao H, Prasada Rao HB, Yang Y, Fong JH, Cloutier JM, Deacon DC, et al. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet. 2014; 46:194–199. PMID:
24390283.

26. Sandor C, Li W, Coppieters W, Druet T, Charlier C, Georges M. Genetic variants in
REC8,
RNF212, and
PRDM9 influence male recombination in cattle. PLoS Genet. 2012; 8:e1002854. PMID:
22844258.
27. Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med. 2008; 359:2143–2153. PMID:
19005198.

28. Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004; 23:1000–1004. PMID:
14647419.

29. Rosell R, Skrzypski M, Jassem E, Taron M, Bartolucci R, Sanchez JJ, et al. BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One. 2007; 2:e1129. PMID:
17987116.

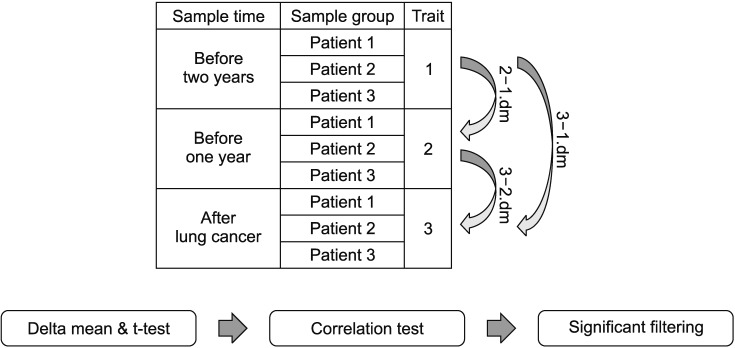
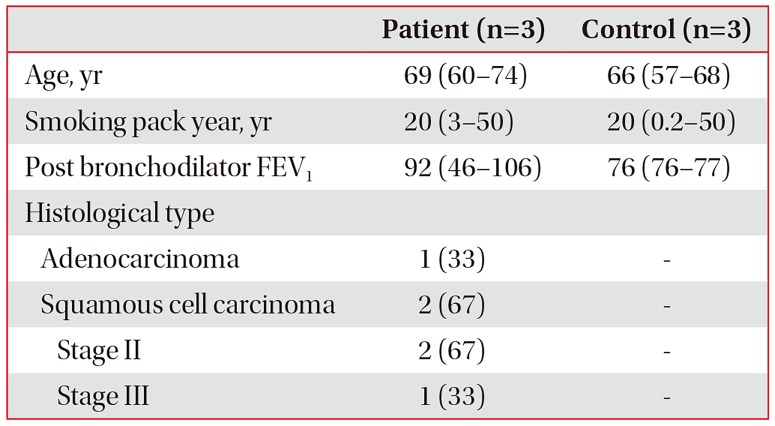




 PDF
PDF ePub
ePub Citation
Citation Print
Print



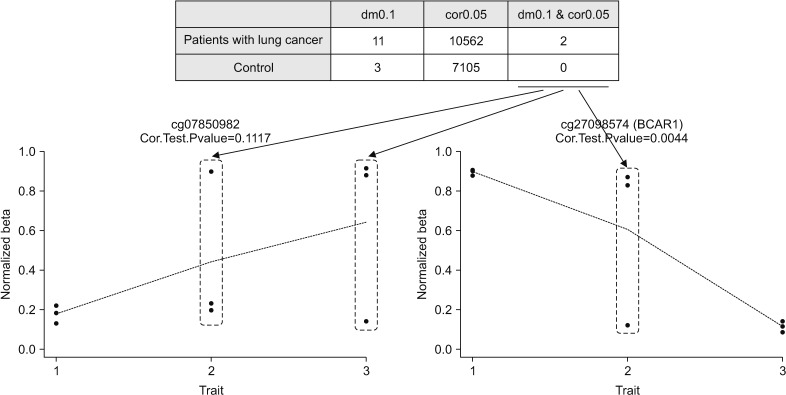
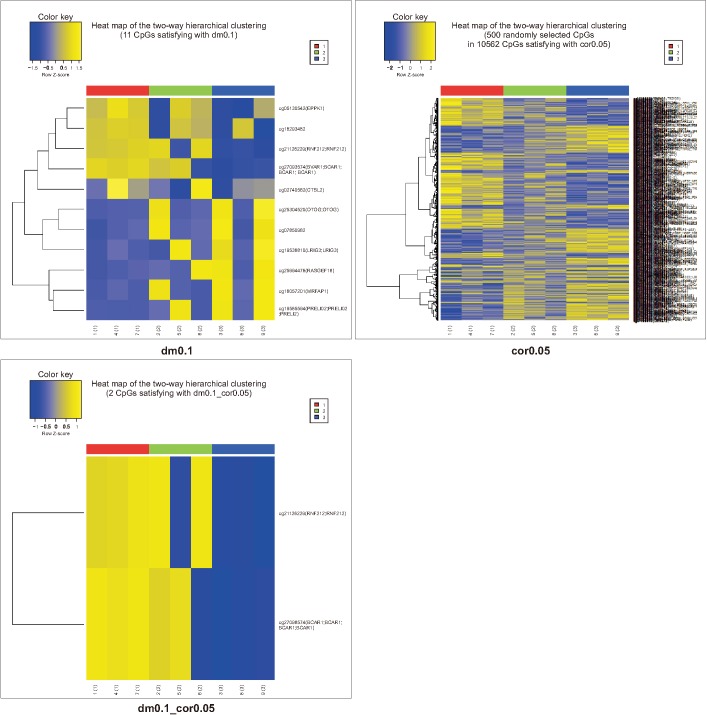
 XML Download
XML Download