INTRODUCTION
METHODS
Cell lines
Plasmids and small interfering RNA
Quantitative real-time PCR
Gene microarray analysis
EdU incorporation assay
MTS assay
Colony formation assay
Western blotting
Immunofluorescence analysis
Co-immunoprecipitation assay
Luciferase reporter assay
Electrophoretic mobility shift assay
Statistical analysis
RESULTS
Pokemon deletion aggravates TGFβ-induced cell proliferation arrest in breast cancer cells
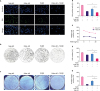 | Figure 1Aggravation of TGFβ-induced cell proliferation arrest after Pokemon deletion in breast cancer cells. After transiently transfected with Neg-siR, Poke-siR (100 nM) or treated with TGFβ1 (10 ng/mL) for 48 hours in MDA-MB-231 cells (A, B), the number of EdU positive cells (EdU+, green color), which represented DNA replication ability, was reduced than control group. (C-E) Alternative siRNA or TGFβ treatment for 48 hours, the viability and proliferation of breast cancer cells were significantly weakened. (F) The colony number of breast cancer cells was also decreased by the 2 treatments. In the above experiments, Pokemon deficiency could promote the growth arrest by TGFβ protein treatment (n = 4).TGFβ = transforming growth factor β; Neg-siR = negative siRNA; Poke-siR = Pokemon siRNA.
*p < 0.01, †p < 0.001.
|
Pokemon modulates TGFβ-Smad4 pathway in breast cancer cells
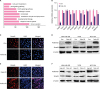 | Figure 2Negative connection between Pokemon and TGFβ-Smad4. (A) Up-regulation of Pokemon affected a series of cell signaling pathway, which was evaluated by the gene array analysis. (B) The mRNA levels of cell invasion-related genes were detected by quantitative real-time polymerase chain reaction after the transfection of Poke-OE or Con. (C) IF assay identified up- or down-regulation of Pokemon reduced or increased expression of Smad4. (D) Western blot results indicated transfection of over-expressed plasmid of Pokemon reduced protein levels of Smad4, and the Pokemon siRNA transfection showed the adverse effects (n = 4).TGFβ = transforming growth factor β; Poke-OE = Pokemon over-expression plasmid with GFP label; Con = negative plasmid; IF= immunofluorescence; MAPK= mitogen-activated protein kinase; DAPI = 4',6-diamidino-2-phenylindole.
*p < 0.05, †p < 0.01.
|
Pokemon regulates the expression of multiple proliferation-associated genes in the TGFβ-Smad4 pathway
 | Figure 3Pokemon regulation on multiple proliferation associated genes. (A-C) Quantitative real-time polymerase chain reaction assay identified that Pokemon knockdown increased TGFβ1 gene level and reduced several proliferation-associated gene levels in MDA-MB-231, T47D and MCF-10A cells (n = 4). (D) Protein levels of CYCLIND1 and CDKN1A were reduced by Pokemon knockdown.TGFβ = transforming growth factor β; Neg-siR = negative siRNA; Poke-siR = Pokemon siRNA; CDKN1A = cyclin dependent kinase inhibitor 1A; CDKN1B = cyclin dependent kinase inhibitor 1B; GADD45B = growth arrest and DNA-damage-inducible, beta; IGFBP3 = insulin-like growth factor-binding protein 3; ATF3 = activating transcription factor 3; CDC6 = cell division cycle 6; EMP1 = epithelial membrane protein 1; PTK2 = protein tyrosine kinase 2; ACVR1 = activin A receptor, type I.
*p < 0.05, †p < 0.01, ‡p < 0.001.
|
Pokemon was bound with SP1 to indirectly suppress Smad4 promoter activity
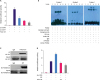 | Figure 4Indirect inhibition of Pokemon on Smad4 promoter activity through binding with SP1. (A) Dual luciferase reporter system showed that transfection of Poke-OE reduced Smad4 luciferase activity (pLuc366 and pLuc207, Smad4 expression plasmid vectors in MDB-MA-231 cells). (B) EMSA assay applied with 2 Smad4 probes proved Pokemon was not bound to promoter active regions of Smad4. (C) Co-immunoprecipitation identified that Pokemon was closely bound to SP1 protein. (D) SP1 promoted luciferase activity of Smad4, which was dose-depend weaken by Pokemon vectors (0.5 µg, 1 µg) in breast cancer cells (n = 3).SP1 = specificity protein 1; Poke-OE = Pokemon over-expressing vectors; IP = immunoprecipitation; IB = immunoblotting; Ab = antibody.
*p < 0.05, †p < 0.01.
|
Pokemon and Smad4 showed contrasting expression levels in different types of breast cancer
 | Figure 5Higher expression level of Pokemon in luminal A type than Basal-like breast cancer tissues, which was just opposite for Smad4. (A, B) The mRNA expression of Pokemon was higher in luminal A type, but lower in basal-like type breast carcinomas, which was just opposite to Smad4. Data was obtained from 2 breast carcinoma database systems.HER2 = human epidermal growth factor receptor 2.
*p < 0.05, †p < 0.01, ‡p < 0.001.
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download