1. Grootjans W, Tixier F, van der, Vriens D, Le Rest CC, Bussink J, et al. The impact of optimal respiratory gating and image noise on evaluation of intratumor heterogeneity on 18F-FDG PET imaging of lung cancer. J Nucl Med. 2016; 57:1692–1698. PMID:
27283931.

2. Yun BL, Cho N, Li M, Jang MH, Park SY, Kang HC, et al. Intratumoral heterogeneity of breast cancer xenograft models: texture analysis of diffusion-weighted MR imaging. Korean J Radiol. 2014; 15:591–604. PMID:
25246820.

3. Bashir U, Siddique MM, Mclean E, Goh V, Cook GJ. Imaging heterogeneity in lung cancer: techniques, applications, and challenges. AJR Am J Roentgenol. 2016; 207:534–543. PMID:
27305342.

4. Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics. 2017; 37:1483–1503. PMID:
28898189.

5. Ganeshan B, Miles KA. Quantifying tumour heterogeneity with CT. Cancer Imaging. 2013; 13:140–149. PMID:
23545171.

6. Miles KA, Ganeshan B, Hayball MP. CT texture analysis using the filtration-histogram method: what do the measurements mean? Cancer Imaging. 2013; 13:400–406. PMID:
24061266.

7. Daginawala N, Li B, Buch K, Yu H, Tischler B, Qureshi MM, et al. Using texture analyses of contrast enhanced CT to assess hepatic fibrosis. Eur J Radiol. 2016; 85:511–517. PMID:
26860661.

8. Lubner MG, Malecki K, Kloke J, Ganeshan B, Pickhardt PJ. Texture analysis of the liver at MDCT for assessing hepatic fibrosis. Abdom Radiol (NY). 2017; 42:2069–2207. PMID:
28314916.

9. Ganeshan B, Abaleke S, Young RC, Chatwin CR, Miles KA. Texture analysis of non-small cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging. 2010; 10:137–143. PMID:
20605762.

10. Chae HD, Park CM, Park SJ, Lee SM, Kim KG, Goo JM. Computerized texture analysis of persistent part-solid ground-glass nodules: differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology. 2014; 273:285–293. PMID:
25102296.

11. Miles KA, Ganeshan B, Griffiths MR, Young RC, Chatwin CR. Colorectal cancer: texture analysis of portal phase hepatic CT images as a potential marker of survival. Radiology. 2009; 250:444–452. PMID:
19164695.

12. Solomon J, Mileto A, Nelson RC, Roy Choudhury K, Samei E. Quantitative features of liver lesions, lung nodules, and renal stones at multi-detector row CT examinations: dependency on radiation dose and reconstruction algorithm. Radiology. 2016; 279:185–194. PMID:
26624973.

13. Yasaka K, Akai H, Mackin D, Court L, Moros E, Ohtomo K, et al. Precision of quantitative computed tomography texture analysis using image filtering: a phantom study for scanner variability. Medicine (Baltimore). 2017; 96:e6993. PMID:
28538408.
14. Mackin D, Fave X, Zhang L, Fried D, Yang J, Taylor B, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015; 50:757–765. PMID:
26115366.

15. Zhao B, Tan Y, Tsai WY, Qi J, Xie C, Lu L, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep. 2016; 6:23428. PMID:
27009765.

16. Berrington de González A, Mahesh M, Kim KP, Bhargavan M, Lewis R, Mettler F, et al. Projected cancer risks from computed tomographic scans performed in the United States in 2007. Arch Intern Med. 2009; 169:2071–2077. PMID:
20008689.

17. Liu L. Model-based iterative reconstruction: a promising algorithm for today’s computed tomography imaging. J Med Imaging Radiat Sci. 2014; 45:131–136.

18. Yu MH, Lee JM, Yoon JH, Baek JH, Han JK, Choi BI, et al. Low tube voltage intermediate tube current liver MDCT: sinogram-affirmed iterative reconstruction algorithm for detection of hypervascular hepatocellular carcinoma. AJR Am J Roentgenol. 2013; 201:23–32. PMID:
23789655.

19. Yoon JH, Lee JM, Yu MH, Baek JH, Jeon JH, Hur BY, et al. Comparison of iterative model-based reconstruction versus conventional filtered back projection and hybrid iterative reconstruction techniques: lesion conspicuity and influence of body size in anthropomorphic liver phantoms. J Comput Assist Tomogr. 2014; 38:859–868. PMID:
25321625.
20. Chang W, Lee JM, Lee K, Yoon JH, Yu MH, Han JK, et al. Assessment of a model-based, iterative reconstruction algorithm (MBIR) regarding image quality and dose reduction in liver computed tomography. Invest Radiol. 2013; 48:598–606. PMID:
23511193.

21. Kim H, Park CM, Lee M, Park SJ, Song YS, Lee JH, et al. Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One. 2016; 11:e0164924. PMID:
27741289.

22. Barrett HH, Myers KJ, Hoeschen C, Kupinski MA, Little MP. Task-based measures of image quality and their relation to radiation dose and patient risk. Phys Med Biol. 2015; 60:R1–R75. PMID:
25564960.

23. Shin JM, Kim TH, Haam S, Han K, Byun MK, Chang YS, et al. The repeatability of computed tomography lung volume measurements: comparisons in healthy subjects, patients with obstructive lung disease, and patients with restrictive lung disease. PLoS One. 2017; 12:e0182849. PMID:
28796825.

24. Geyer LL, Schoepf UJ, Meinel FG, Nance JW Jr, Bastarrika G, Leipsic JA, et al. State of the Art: Iterative CT Reconstruction Techniques. Radiology. 2015; 276:339–357. PMID:
26203706.

25. Willemink MJ, de Jong PA, Leiner T, de Heer LM, Nievelstein RA, Budde RP, et al. Iterative reconstruction techniques for computed tomography Part 1: technical principles. Eur Radiol. 2013; 23:1623–1631. PMID:
23314600.

26. Ng F, Ganeshan B, Kozarski R, Miles KA, Goh V. Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology. 2013; 266:177–184. PMID:
23151829.

27. Elpek GÖ. Angiogenesis and liver fibrosis. World J Hepatol. 2015; 7:377–391. PMID:
25848465.

28. Lautt WW. Mechanism and role of intrinsic regulation of hepatic arterial blood flow: hepatic arterial buffer response. Am J Physiol. 1985; 249(5 Pt 1):G549–G556. PMID:
3904482.

29. Yoon JH, Lee JM, Klotz E, Jeon JH, Lee KB, Han JK, et al. Estimation of hepatic extracellular volume fraction using multiphasic liver computed tomography for hepatic fibrosis grading. Invest Radiol. 2015; 50:290–296. PMID:
25493416.

30. Park SB, Kim YS, Lee JB, Park HJ. Knowledge-based iterative model reconstruction (IMR) algorithm in ultralow-dose CT for evaluation of urolithiasis: evaluation of radiation dose reduction, image quality, and diagnostic performance. Abdom Imaging. 2015; 40:3137–3146. PMID:
26197735.

31. Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015; 5:11075. PMID:
26242464.

32. Shafiq-Ul-Hassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci Rep. 2018; 8:10545. PMID:
30002441.

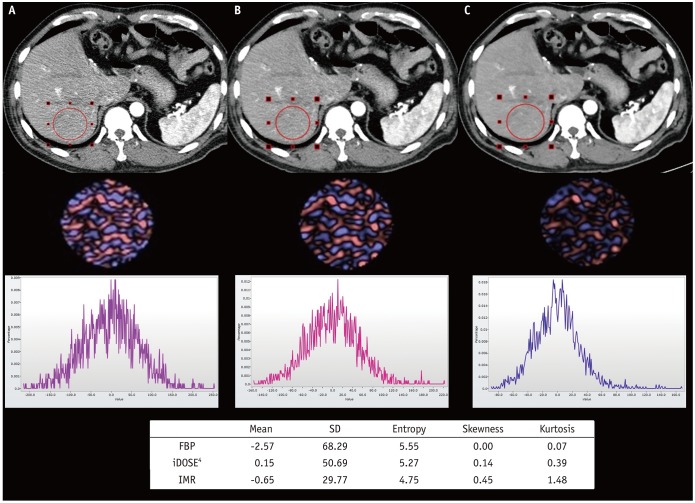
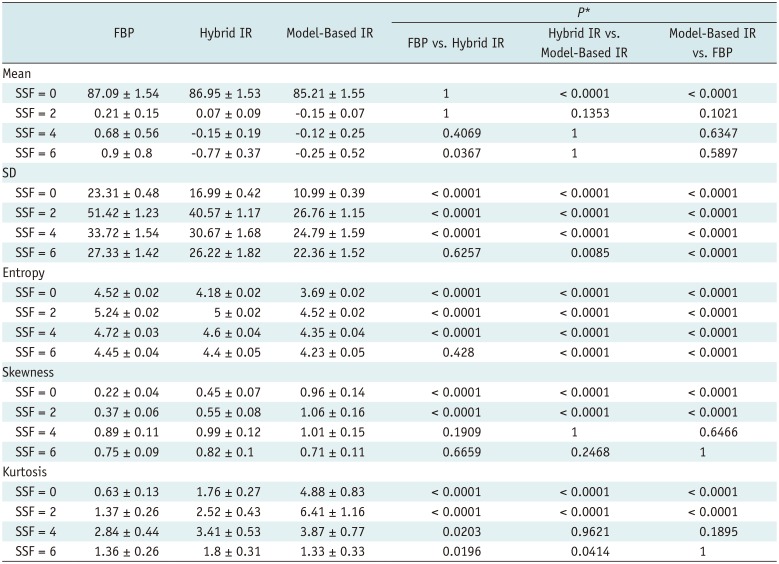
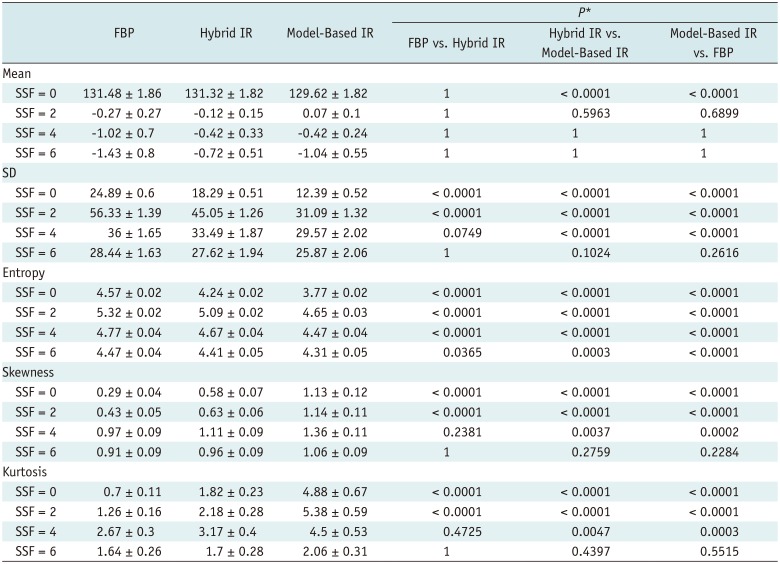
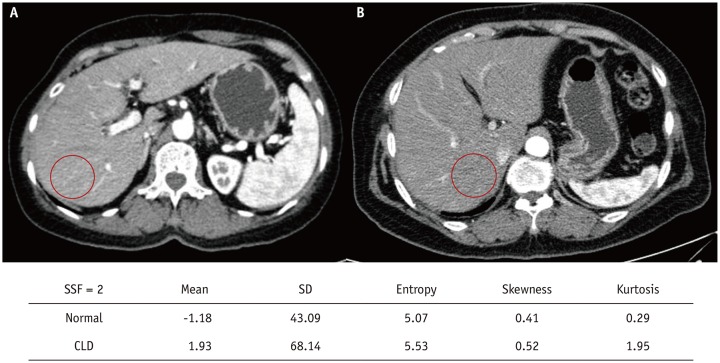
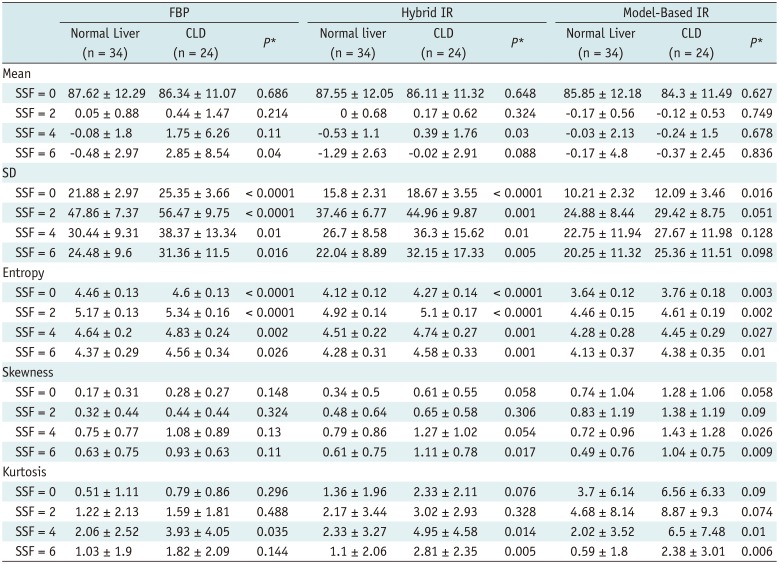
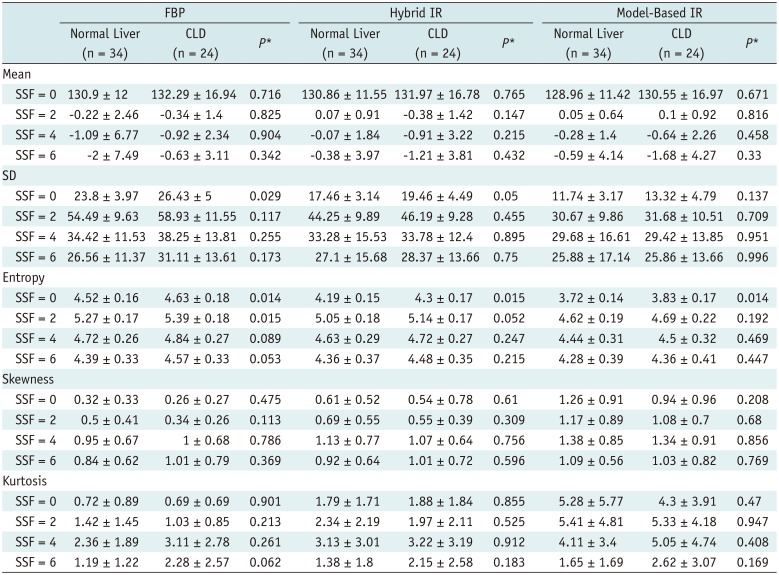
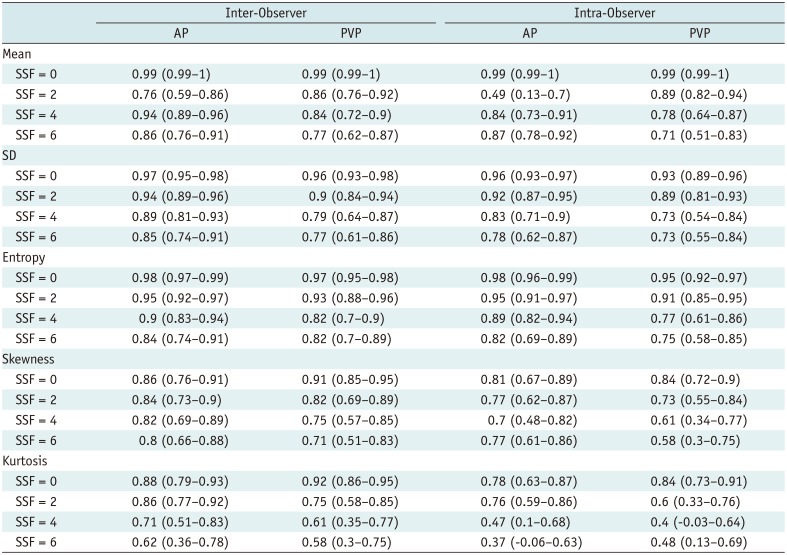




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download