This article has been
cited by other articles in ScienceCentral.
Dear Editor,
The
SET-NUP214 fusion gene has been observed most commonly in T-cell ALL (T-ALL) patients, with a frequency of about 3.3% to 10.3%, and in acute undifferentiated leukemia (AUL) patients, but is rare in AML patients [
1234]. Most patients diagnosed as having T-ALL have normal karyotypes according to conventional cytogenetic analysis [
2].
SET-NUP214 fusion, though reported rarely, has a significant value in acute leukemia because it often predicts poor patient outcomes [
1]. We report a rare case of AML with
SET-NUP214 fusion and massive hyperdiploidy. To the best of our knowledge, this is the first case of AML with
SET-NUP214 fusion in Korea. This study was approved by the Institutional Review Board of Dong-A University Hospital, Busan, Korea, and informed consent was obtained from the patient.
A 46-year-old Korean male visited the emergency department of Dong-A University Hospital for further evaluation of fever and general weakness. Clinical evaluations were unremarkable, except for mild haziness on a chest X-ray, suggesting pneumonia. Initial laboratory results showed mild leukocytosis (17.1×10
9/L leukocytes), severe anemia (53 g/L Hb), and thrombocytopenia (120×10
9/L platelets). Differential count revealed 89% blasts with 1% segmented neutrophils, 7% lymphocytes, and 3% monocytes. A bone marrow aspirate smear showed 88% of pleomorphic blasts characterized by large size, high nucleic/cytoplasmic ratio, irregular nuclear membranes and convolutions, and cytoplasmic projections (
Fig. 1A). A bone marrow biopsy revealed portions of marked hypercellular areas and moderate fibrosis. The blasts were negative for periodic acid-Schiff and non-specific esterase stains and mostly positive for Sudan black B and myeloperoxidase (MPO) stains. In flow-cytometric analysis, the blasts were positive for MPO, CD33, CD7, CD34, and CD71 antigens, indicating AML.
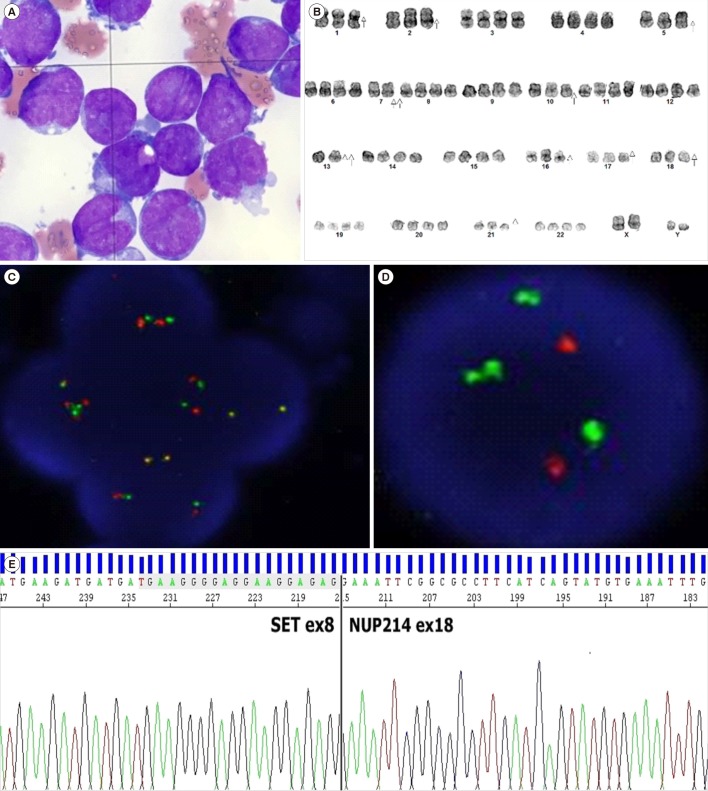 | Fig. 1Features of bone marrow aspirate smear and results of chromosomal analysis, FISH, and Sanger sequencing. (A) Bone marrow aspirate smear showing leukemic blasts comprising mostly myeloblasts (Wright-Giemsa stain, ×1,000). (B) Chromosomal analysis showing 59–90,XXYY,−1,−2,−5,−7,−7,−10,−13,−13,−16,−17,−18,−21[cp23]. (C) FISH analysis with a SET/NUP214 probe showing two yellow fusions, and two green and two red probes, suggesting either del(9)(q34.11q34.13) or t(9;9)(q34;q34). (D) FISH analysis with ABL1 probe showing two red (ABL1) and four green (BCR) signals. (E) Sanger sequence analysis of SET and NUP241.
|
Multiplex-nested PCR with HemaVision (DNA Diagnostic A/S, Risskov, Denmark) revealed
SET-NUP214 fusion transcripts of 393 base pairs. Chromosome analysis revealed 59–90,XXXY,−1, −2,−5,−7,−7,−10,−13,−13,−16,−17,−18,−21[cp23], interpreted as a composite karyotype with massive hyperdiploidy in all of the 23 examined chromosomes, without any apparent structural abnormalities (
Fig. 1B). FISH showed tetrasomy signals for the
MLL,
ERG1,
CEP7,
TP53, and
MECOM genes. Because
SET-NUP214 rearrangement can result from either del(9)(q34.11q34.13) or t(9;9)(q34;q34) [
5], further studies with additional FISH probes were conducted. Based on the results of FISH analysis with a
BCR/ABL1 probe, we suspected that the
SET-NUP214 fusion gene in this patient was probably formed by deletion of chromosome 9, because
ABL1 (9q34.12) is located between
SET (9q34.11) and
NUP214 (9q34.13), and the two red signals were missing (
Fig. 1C and 1D) [
67]. However, Sanger sequencing indicated that the
SET-NUP214 fusion was generated by chromosomal translocation (
Fig. 1E).
The patient was diagnosed as having AML without maturation with SET-NUP214 fusion. Induction and consolidation chemotherapy with idarubicin and cytosine arabinoside was given, and in approximately three months, the patient achieved complete hematologic (2.6% of myeloblasts) and molecular remissions (normal male karyotype and negative for SET-NUP214 fusion as indicated by PCR). After approximately five months, the patient received allogenic peripheral blood stem cell transplantation from a full-matched sibling donor. Currently, with successful platelet and neutrophil engraftment and sufficient CD34 counts, the patient is being treated for cytomegalovirus infection.
Since
SET-NUP214 fusion was first detected in an AML patient with a normal karyotype in 2007 [
2], 43 cases of acute leukemia with such rearrangement, including the current case, have been reported worldwide [
1]. AML comprises only a very small portion (~2.4%) of these cases, and a hyperdiploid karyotype is also extremely rare. Characteristics of a previous [
8] and the present case of AML with
SET-NUP214 fusion are presented in
Table 1.
Table 1
Characteristics of a previous and the present case of AML with SET-NUP214 fusion

|
No. case (N) |
Sex |
Age (year) |
Dx. |
Additional chromosomal abnormalities |
|
Reference |
|
1 |
M |
35 |
AML-M4 |
None |
del(9)(q34.11q34.13) |
Rosati et al. [8] |
|
2 |
M |
46 |
AML-M1 |
59–90,XXXY,-1,-2,-5,-7,-7,-10,-13,-13,-16,-17,-18,-21[cp23] |
t(9;9)(q34;q34) |
Present case |

NUP214 is essential for cell cycle progression and nucleocytoplasmic transport, and
SET is involved in inhibiting apoptosis caused by cytotoxic T lymphocytes [
9]. The fusion of these genes is recurrent in T-ALL, and it is strongly associated with corticosteroid and chemotherapy resistance [
10]. However, the prognostic significance of this fusion in AML with numerical chromosomal abnormalities remains unclear. We believe that such genetic rearrangements with additional chromosomes are clinically heterogeneous, and it is difficult to place the current case into a specific prognostic group. Because this genetic rearrangement is cryptic in conventional karyotyping, additional analytical methods, such as reverse transcription PCR or FISH, should be used.






 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download