Abstract
Background
Human astroviruses (HAstVs) are one of the major causes of acute gastroenteritis. Classic HAstVs can be classified into eight genotypes. We investigated the positive rate of HAstVs and the distribution of HAstV genotypes in strains isolated from patients with acute gastroenteritis in Hwaseong, Korea, in 2013–2017.
Methods
Between November 2013 and December 2017, 3,519 stool samples were collected from patients with symptoms of acute gastroenteritis and tested for HAstV using multiplex PCR. For HAstV-positive samples, the ORF2 gene, which encodes a capsid protein, was genotyped by reverse-transcription PCR and sequencing. Phylogenetic analysis was performed to determine whether the sequences of the HAstVs differed by year.
Results
The positive rate of HAstV was 1.9% (67 samples). HAstV Type 1 was the most prevalent genotype (82.4%), and Types 4, 5, and 8 were also detected. Infection occurred year-round, with no distinct seasonal variation. Infection occurred at nearly all ages (55 days–81 years; median: 3 years), and the positive rate was substantially higher in children younger than five years. Phylogenetic analysis revealed three distinct clades of HAstV Type 1 according to the collection time.
Conclusions
Our results provide recent epidemiological data on HAstVs in Korea between 2013 and 2017. The finding of three distinct clades of HAstV Type 1 according to collection time suggests genetic evolution of HAstVs. These findings can enhance our knowledge on HAstV infection and viral evolution.
Human astroviruses (HAstVs) are enteric pathogens that cause acute gastroenteritis in young children and immunocompromised/elderly people. The prevalence of classic HAstV infection ranges from 2% to 9% in children with diarrhea, although incidences of 61% have been reported [123456]. They are non-enveloped, single-stranded, positive-sense RNA viruses of approximately 6.8 kb in length that contain three overlapping open reading frames (ORFs): ORF1a, ORF1b, and ORF2. ORF1a and OFR1b encode the non-structural viral proteins, including RNA-dependent RNA polymerase, and ORF2 encodes the capsid protein precursor that is cleaved into capsid proteins VP34, VP27, and VP25 [1]. Sequence variation among astroviruses is the greatest in the ORF2 region, which encodes a highly conserved N-terminal domain (aa 1–424), a hypervariable domain (HVR) (aa 425–688), and a C-terminal domain [1].
Classic HAstVs (Mamastrovirus 1) have been classified into eight genotypes on the basis of capsid antigenicity. HAstV Type 1 is the most prevalent genotype, whereas Types 2–8 are less prevalent [1235]. HAstV Type 2 was the most prevalent genotype in Sao Paulo, Brazil, in 1994–1996, and in Mexico, in 1988–1991 [78], and HAstV Type 5 was the most prevalent genotype in Linyi, China, in 2013 [9]. Therefore, the genotypic distribution of HAstVs may vary geographically and temporally [1]. In Korea, several studies have examined the positive rate and genotype of HAstVs, but they have been conducted either several years ago or for very short durations [1011121314]. We investigated the positive rate of HAstVs and the distribution of HastV genotypes in Hwaseong, a city with high population density in Korea, in 2013–2017 and examined the sequence variations of HAstVs.
Between November 2013 and December 2017, 3,519 stool samples from patients with acute gastroenteritis were tested for diarrhea-causing viruses in the laboratory of Hallym University Dongtan Sacred Heart Hospital, Hwaseong, Korea. After the test, residual positive stool samples were routinely collected and stored at −70℃ until use. This study was retrospectively performed on stored HAstV-positive stool samples. The study protocol was approved by the Institutional Review Board (IRB) of Hallym University Dongtan Sacred Heart Hospital (IRB No. 2014-069, NON-2018-003), with waivers of informed consent. Data regarding the patients' ages and symptoms were collected by reviewing patients' medical records.
Stool samples were diluted to a 10% suspension in phosphate-buffered saline, and viral RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) and the QIAcube platform (Qiagen). HAstV was detected using Seeplex Diarrhea-V ACE Detection assays (Seegene, Seoul, Korea), which employ a multiplex PCR technique for detecting rotavirus, norovirus, adenovirus, and HAstV simultaneously, and a PTC-200 Thermal Cycler (Bio-Rad, Hercules, CA, USA). All procedures were performed according to the manufacturers' instructions. After PCR, residual positive samples were collected and stored at −70℃ for further study.
Genotyping was performed by reverse-transcription (RT) PCR and sequencing in January 2018. HAstV PCR-positive stored stool samples were thawed, and viral RNA was extracted again from these samples. cDNA was generated from RNA using the Invitrogen SuperScript III First Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA) and used for PCR with DNA AmpliTaq Gold Taq (Applied Biosystems, GmbH, Weiterstadt, Germany). We used a gene-specific primer set, PreCAP1 (5′-GGACTGCAAAGCAGCTTCGTG-3′) and 82b (5′-GTGAGCCACCAGCCATCCCT-3′), which targets the ORF2 gene and generates a 719-bp PCR product [15]. For samples that did not yield a PCR product using these primers, multiplex PCR was conducted using a different primer set that could amplify all classic serotypes of HAstVs [16]. A positive control (RNA extracted from the sample of an HAstV-positive patient) and a negative control (RNase-free water) were co-amplified to assess false-negative and false-positive results, respectively. PCR products were visualized after agarose gel electrophoresis, purified using the QIAquick PCR Purification Kit (Qiagen), and sequenced using ABI BigDye Terminator v3.1 Cycle Sequencing Kits (Applied Biosystems Inc., Foster City, CA, USA) on the 3500XL Genetic Analyzer (Thermo Fisher Scientific). HAstV genotypes of the obtained sequences were determined by BLAST searches against the GenBank database.
Phylogenetic analysis was conducted to evaluate genetic relationships between the HAstVs obtained and the closest strains in GenBank. The accession numbers of the closest strains were KM981712.1, MG932591.1, JQ898572.1, KP453847.2, KM981734.1, MG932580.1, KF420152.1, MG932569.1, KP862744.1, and NC_001943.1. The HAstV sequences were aligned using MEGA version 7 [17]. Phylogenetic trees were constructed using the maximum-likelihood method and Tamura-Nei substitution models with 1,000 bootstrap replications, using MEGA version 7.
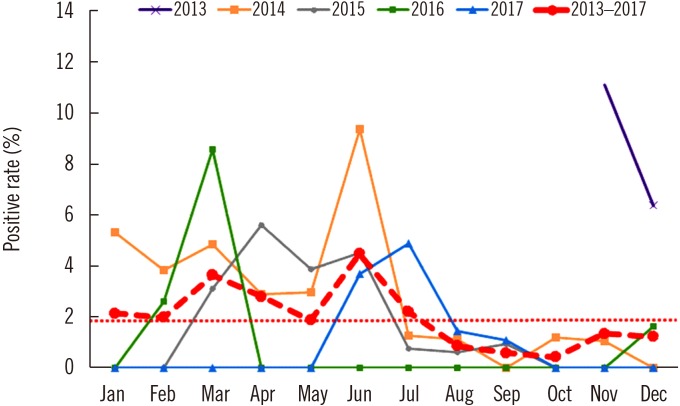
HAstV positive rates according to patient age are shown in Table 1. The average positive rate between 2013 and 2017 was 1.9%, and the ages of HAstV-positive patients ranged from 55 days to 81 years (median, 3 years, and average, 15.7 years). Prevalence was higher in children under five years; however, 26 out of 67 patients (38.8%) were over five years. Table 2 shows the major symptoms of HAstV infection based on the patients' medical records. All patients showed at least one of the symptoms: diarrhea, nausea or vomiting, abdominal or epigastric pain, and fever. Monthly positive rates are shown in Fig. 1. The highest positive rates in each year were observed during November in 2013 (11.1%, 3/27), June in 2014 (9.4%, 6/64), April in 2015 (5.6%, 6/107), March in 2016 (8.6%, 3/35), and July in 2017 (4.9%, 7/143). In 24 of the total 50 months considered, HAstV was not detected.
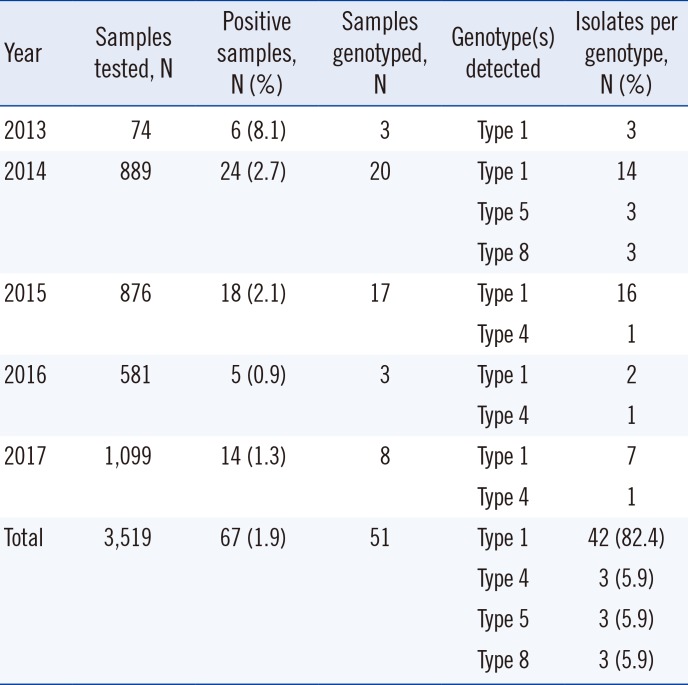
Of 67 HAstV-positive samples, 51 were successfully genotyped (Table 3). Eight samples were discarded after PCR by mistake, and eight samples yielded no PCR product with the genotyping primer sets used in this study. HAstV Type 1 was the most prevalent (82.4%), and Types 4, 5, and 8 were also detected. Types 5 and 8 were detected only in 2014, whereas Type 4 was detected in 2015, 2016, and 2017 (Table 3).
Phylogenetic analysis is shown in Fig. 2. There were three distinct clades among HAstV Type 1 according to three time periods (2013–2014, 2014–2016, and 2017). The five strains detected in 2017 (D-1389, D-1397, D-1413, D-1415, and D-1421) were phylogenetically closest to an HAstV strain detected in Russia in 2017 (MG932591.1). The four strains detected in 2013 and 2014 (D-34, D-36, D-343, and D-423) were phylogenetically closest to a strain detected in Thailand in 2011 (JQ898572.1), and most strains detected during 2014–2016 were phylogenetically closest to a strain detected in China in 2013 (KM981712.1). The three strains of HAstV Type 4 (D-856, D-1153, and D-1403) were phylogenetically closest to a strain detected in Russia in 2017 (MG932580.1) and one detected in China in 2013 (KM981734.1). The three strains of HAstV Type 5 (D-52, D-233, and D-322) were closest to a strain detected in Bangladesh in 2012 (KF420152.1) and one detected in Russia in 2012 (MG932569.1). The three strains of HAstV Type 8 (D-123, D-230, and D-269) were the closest to a strain detected in Korea in 2014 (KP862744.1).
The overall HAstV positive rate in this study was 1.9%. The positive rate of HAstV differs across regions [1] from 0.21% in Chongquing, China [18] to 61% in Mayan infants [19], while the average incidence worldwide is 11% [1]. The positive rate of HAstV can differ with respect to time and has tended to decrease in the last decade [1]. In Korea, the HAstV positive rate was less than 2% [10111213], and only one study reported a positive rate up to 4% in Cheonan [14]. The present study confirmed that the positive rate of HAstV in Korea in recent years is still lower than the average global positive rate of 11% [1]. This may be due to the current socio-economic and sanitary status of Korea [20].
HAstVs are considered to cause infections mainly in children younger than five years [1]. However, in this study, 38.8% of HAstV-positive patients were older than five years, which suggests that HAstV infection may also occur in adolescents and adults. The higher percentage of patients over five years in this study could be attributed to the fact that we collected stool samples from patients of all ages with diarrhea symptoms.
Analysis of the monthly positive rate by year did not show distinct seasonal variations in HAstV infection. Cumulative positive rates revealed a relatively high HAstV incidence from March to July and a relatively low incidence from August to October; however, definitive conclusions cannot be drawn because of the small number of positive samples.
Although HAstV Type 1 was the most prevalent (82.4%), we also detected Types 4, 5, and 8. With a few exceptions, HAstV Type 1 is the most common worldwide [1]. Type 4 was the second most prevalent genotype in Argentina (25%, 1995–1998 [21]), Spain (26%, 1997–2000 [22]), and Australia (15%, 1995 [23]). Type 8 has been detected at a relatively higher frequency in India (16%, 2004–2008 [24]) and Spain (11%, 1997–2000 [22]) than in previous research [1]. Type 5 has been detected at high frequency in Egypt (15.7%, 1995) [125]. We confirmed that Types 5 and 8 strains were isolated only in 2014 and were not small outbreaks or family transmissions, by reviewing patients' medical records. HAstV Types 4, 5, and 8 have been reported in Korea [1013]. Phylogenic analysis showed that the sequences of HAstV Type 1 isolated in this study between November 2013 and August 2017 could be grouped into three clades related to three periods, which may suggest genetic evolution of HAstVs over this period in Korea. This is the first study to analyze a phylogeny of HAstVs according to isolation time in Korea.
One limitation of this study was that only samples that tested positive with the commercially available Seeplex PCR assay were genotyped; thus, types that did not test positive with this assay were probably not detected. The company claims that this PCR assay can detect HAstV Types 1–8, but this assay may not detect HAstVs whose genomes have variations at primer-binding sites. If all the 3,519 samples in this study had been genotyped without the PCR screening procedure, the positive rate of HAstV might have been higher and a greater variety of genotypes may have been detected, because HAstV detection in stool samples might differ according to the detection method and primer pair used.
In conclusion, our results provide recent data on positive rates, genotypes, and sequence variations of HAstVs in Korea between 2013 and 2017. The finding of three distinct clades of HAstV Type 1 according to collection time suggests the genetic evolution of HAstVs. These findings can enhance our knowledge on HAstV infection and viral evolution.
Acknowledgment
This study was supported by the Technology Innovation Program (10047748) funded by the Ministry of Trade, Industry & Energy and by Hallym University Research Fund 2017 (HURF-2017-30).
References
1. Bosch A, Pintó RM, Guix S. Human astroviruses. Clin Microbiol Rev. 2014; 27:1048–1074. PMID: 25278582.
2. De Grazia S, Bonura F, Bányai K, Gellért A, Marineo S, Martella V, et al. Temporal variation in the distribution of type-1 human astrovirus lineages in a settled population over 14 years. Arch Virol. 2016; 161:1633–1637. PMID: 26923926.
3. Lekana-Douki SE, Kombila-Koumavor C, Nkoghe D, Drosten C, Drexler JF, Leroy EM. Molecular epidemiology of enteric viruses and genotyping of rotavirus A, adenovirus and astrovirus among children under 5 years old in Gabon. Int J Infect Dis. 2015; 34:90–95. PMID: 25796432.
4. Monastiri A, Aouni M, Guix S, Mechri B, Lopez-Roig M, Abid NBS, et al. Clinical surveillance for human astrovirus in Monastir region, Tunisia. BMC Public Health. 2016; 16:57. PMID: 26796330.
5. Nakamura N, Kobayashi S, Minagawa H, Matsushita T, Sugiura W, Iwatani Y. Molecular epidemiology of enteric viruses in patients with acute gastroenteritis in Aichi Prefecture, Japan, 2008/09–2013/14. J Med Virol. 2016; 88:1180–1186. PMID: 26647761.
6. Zaraket H, Abou-El-Hassan H, Kreidieh K, Soudani N, Ali Z, Hammadi M, et al. Characterization of astrovirus-associated gastroenteritis in hospitalized children under five years of age. Infect Genet Evol. 2017; 53:94–99. PMID: 28536072.
7. Resque HR, Munford V, Castilho JG, Schmich H, Caruzo TA, Rácz ML. Molecular characterization of astrovirus in stool samples from children in Sao Paulo, Brazil. Mem Inst Oswaldo Cruz. 2007; 102:969–974. PMID: 18209936.
8. Guerrero ML, Noel JS, Mitchell DK, Calva JJ, Morrow AL, Martínez J, et al. A prospective study of astrovirus diarrhea of infancy in Mexico City. Pediatr Infect Dis J. 1998; 17:723–727. PMID: 9726348.
9. Zhou N, Lin X, Wang S, Wang H, Li W, Tao Z, et al. Environmental surveillance for human astrovirus in Shandong Province, China in 2013. Sci Rep. 2014; 4:7539. PMID: 25519005.
10. Jeong AY, Jeong HS, Jo MY, Jung SY, Lee MS, Lee JS, et al. Molecular epidemiology and genetic diversity of human astrovirus in South Korea from 2002 to 2007. Clin Microbiol Infect. 2011; 17:404–408. PMID: 20491833.
11. Kang YH, Park YK, Ahn JB, Yeun JD, Jee YM. Identification of human astrovirus infections from stool samples with diarrhea in Korea. Arch Virol. 2002; 147:1821–1827. PMID: 12209320.
12. Ham H, Oh S, Jang J, Jo S, Choi S, Pak S. Prevalence of human astrovirus in patients with acute gastroenteritis. Ann Lab Med. 2014; 34:145–147. PMID: 24624351.
13. Koo HS, Jo HC, Baik HS. Epidemiological study of outbreak of gastroenteritis associated with Norovirus and astrovirus in Busan, Korea. J Life Sci. 2016; 26:999–1006.
14. Kim JK, Kim JW. Molecular epidemiologic trends of diarrhea-causing virus infection from clinical specimens in Cheonan, Korea, in 2010–2012. J Clin Lab Anal. 2014; 28:47–51. PMID: 24375926.
15. Yan H, Yagyu F, Okitsu S, Nishio O, Ushijima H. Detection of norovirus (GI, GII), sapovirus and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J Virol Methods. 2003; 114:37–44. PMID: 14599677.
16. Sakamoto T, Negishi H, Wang QH, Akihara S, Kim B, Nishimura S, et al. Molecular epidemiology of astroviruses in Japan from 1995 to 1998 by reverse transcription-polymerase chain reaction with serotype-specific primers (1 to 8). J Med Virol. 2000; 61:326–331. PMID: 10861640.
17. Kumar S, Stecher G, Mega TK. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1874. PMID: 27004904.
18. Ren Z, Kong Y, Wang J, Wang Q, Huang A, Xu H. Etiological study of enteric viruses and the genetic diversity of norovirus, sapovirus, adenovirus, and astrovirus in children with diarrhea in Chongqing, China. BMC Infect Dis. 2013; 13:412. PMID: 24004442.
19. Maldonado Y, Cantwell M, Old M, Hill D, Sanchez ML, Logan L, et al. Population-based prevalence of symptomatic and asymptomatic astrovirus infection in rural Mayan infants. J Infect Dis. 1998; 178:334–339. PMID: 9697712.
20. Troeger C, Khalil IA, Rao PC, Cao S, Blacker BF, Ahmed T, et al. Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatr. 2018; 172:958–965. PMID: 30105384.
21. Espul C, Martínez N, Noel JS, Cuello H, Abrile C, Grucci S, et al. Prevalence and characterization of astroviruses in Argentinean children with acute gastroenteritis. J Med Virol. 2004; 72:75–82. PMID: 14635014.
22. Guix S, Caballero S, Villena C, Bartolomé R, Latorre C, Rabella N, et al. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J Clin Microbiol. 2002; 40:133–139. PMID: 11773106.
23. Palombo EA, Bishop RF. Annual incidence, serotype distribution, and genetic diversity of human astrovirus isolates from hospitalized children in Melbourne, Australia. J Clin Microbiol. 1996; 34:1750–1753. PMID: 8784582.
24. Verma H, Chitambar SD, Gopalkrishna V. Astrovirus. Astrovirus associated acute gastroenteritis in western India: predominance of dual serotype strains. Infect Genet Evol. 2010; 10:575–579. PMID: 20117249.
25. Naficy AB, Rao MR, Holmes JL, Abu-Elyazeed R, Savarino SJ, Wierzba TF, et al. Astrovirus diarrhea in Egyptian children. J Infect Dis. 2000; 182:685–690. PMID: 10950760.
Fig. 2
Phylogenetic analysis of HAstVs in this study and reference strains in GenBank. Phylogenetic trees were constructed using the maximum-likelihood method with 1,000 bootstrap replications, using MEGA version 7. The percentage of trees, in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Black circles indicate the HAstV isolates from this study, white squares indicate the reference species from GenBank, and strains are described in terms of accession number, type of HAstV, country of collection, and year of collection.
Abbreviation: HAstV, human astrovirus.

Table 1
Age distribution of HAstV-positive patients
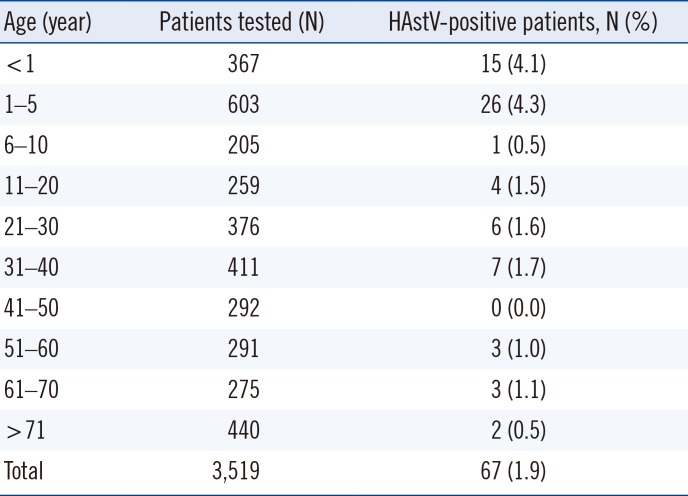
Table 2
Symptoms of HAstV-positive patients (N=67)
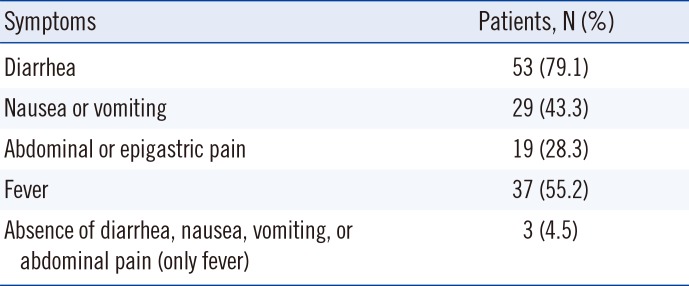
Table 3
Yearly positive rates of HAstV as determined by PCR and genotypes of HAstVs




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download