This article has been corrected. See "Erratum to “MicroRNA-370 Regulates Cellepithelial-Mesenchymal Transition, Migration, Invasion, and Prognosis of Hepatocellular Carcinoma by Targeting GUCD1” by He Y and He X (Yonsei Med J 2019;60:267–276.)" in Volume 60 on page 402.
Abstract
Purpose
Hepatocellular carcinoma (HCC) is a highly aggressive malignant tumor, the prognosis of which remains poor. Recently, microRNAs have been reported to play crucial functions in multiple tumors, including HCC. However, the molecular mechanisms of miR-370 in HCC still remain largely unknown. The present study focused on the effects of miR-370 on HCC migration, invasion, and epithelial-mesenchymal transition (EMT).
Materials and Methods
We investigated the key roles and possible regulatory mechanism of miR-370 in regulating HCC metastasis with functional assays, such as transwell assay. Quantitative real-time PCR (qRT-PCR) was used to detect miR-370 and guanylylcyclase domain containing 1 (GUCD1) expression in HCC tissues and cells. Subsequently, we performed transwell assays to determine the functions of miR-370 in HCC cell invasion and migration. Western blot was used to determine protein expressions of relevant genes. Luciferase reporter assays were conducted to confirm the target gene of miR-370.
Results
qRT-PCR analysis demonstrated that miR-370 was dramatically downregulated in HCC. Moreover, downregulated miR-370 was found to be associated with poor survival and adverse clinicopathologic characteristics of HCC patients. Transwell assays revealed that miR-370 overexpression dramatically suppressed HCC invasion and migration. Meanwhile, miR-370 restoration prominently inhibited EMT progression in HCC cells. Luciferase reporter assays confirmed GUCD1 as a downstream target gene of miR-370. GUCD1 expression in HCC tissues was prominently increased and inversely correlated with miR-370 expression. Furthermore, GUCD1 was verified as mediating the suppressive influence of miR-370 on cell metastasis and EMT in HCC.
Hepatocellular carcinoma (HCC) is one of the most common and fatal liver cancers, with high morbidity and mortality rates all over the world.12 Although surgical resection, percutaneous ethanol injection, and liver transplantation have improved survival of HCC, the prognosis of HCC patients remains less than ideal because of poor responses to chemotherapy and radiotherapy, intrahepatic and extrahepatic metastasis, and recurrence.3 Delineating molecular signatures of metastatic potential and risk of recurrence in HCC may allow for development of therapies that can be applied earlier in HCC. Although several biomarkers have recently been reported,45 there still remains a lack of appropriate predictors that can be widely applied in the clinical setting of HCC.6
Epithelial-mesenchymal transition (EMT) is a notable biological event in the process of cancer-related migration and invasion, exhibiting marked morphological changes between epithelial state and mesenchymal cell-like properties.7 EMT is characterized by the loss of cell-cell contact through the inhibition of epithelial markers, such as E-cadherin expression, and the acquisition of mesenchymal features, such as the upregulation of the mesenchymal markers N-cadherin and vimentin. Tumor cells are endowed with invasive and migratory properties that allow them to migrate through the extracellular matrix to distant organs during EMT. Some research has shown that EMT plays crucial roles in the progression of different tumors, including lung carcinoma,8 breast carcinoma,9 and bladder carcinoma.10 Further investigations are needed to better understand the mechanism underlying EMT progression in HCC.
MicroRNAs (miRNAs) are a group of single-stranded, small, endogenous, non-coding RNAs that act as important mediators of basic biological processes by binding to complementary sequences in 3′UTR of target mRNAs.1112 Additionally, it has recently been reported that miRNAs may serve as tumor suppressors or oncogenes during the progression and development of tumors, indicating the potential of miRNAs to be biomarkers for tumor diagnosis and therapy.13 For example, Hu, et al.14 reported that miR-375 suppressed cell growth and invasion in esophageal carcinoma by inhibiting metadherin expression; Guan, et al.15 found that miR-93 promoted gastric cancer cell proliferation and metastasis by regulating TIMP2; miR-222 was found to influence colorectal carcinoma cell invasion and migration by targeting MIA3.16 Here, we focused our attention on miR-370, a novel cancer-related miRNA, which has been found to be dysregulated in cancers, including HCC.17 In a previous study, miR-370 was reported as exerting anti-tumor functions in HCC by regulating PIM1.18 We speculated that miR-370 may participate in HCC EMT and metastasis via the regulation of a specific target gene.
Given the pivotal roles of miR-370 in HCC progression, identification of key candidate targets could be essential for the investigation of underlying molecular mechanisms. Guanylylcyclase domain containing 1 (GUCD1) is a ubiquitously expressed and highly conserved gene, which was found to be significantly upregulated in HCC and to play important roles in liver tumorigenesis.19 Previous study showed that hepatic cyclic adenosine monophosphate (cAMP) concentrations increased during liver regeneration and that cAMP was responsible for the phosphorylation and activation of many cAMP-regulated transcription factors.20 Analysis of GUCD1 mRNA levels showed that its transcription decreased following stimulation with cAMP.21 Therefore, high expression of GUCD1 mRNA in many cell lines derived from human cancers prompted us to hypothesize a well-defined role in the mechanisms regulating HCC progression. Nevertheless, the regulatory functions of GUCD1 in HCC remain largely unclear. In this study, we measured GUCD1 expression and investigated correlations between miR-370 and GUCD1 in HCC.
Tissue specimens were collected from 50 HCC patients who underwent surgical resection at Taixing People's Hospital between February 2015 and July 2017. No patient had received chemotherapy or radiotherapy prior to the surgery. All surgical samples were snap-frozen in liquid nitrogen and stored at −80℃ for further use. The experiments were approved by the Ethics Committee of Taixing People's Hospital. All HCC patients involved in the current study provided written informed consent.
Human HCC cells (Huh7, Bel-7402, HCCLM3, Hep3B, and SMMC-7721) and normal liver cells LO2 were purchased from the Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Gibco, Grand Island, NY, USA) with 5% CO2 at 37℃.
All miRNA-related vectors, such as miR-370 mimics, inhibitor, or miR-control were synthesized by GenePharma (Shanghai, China). The GUCD1expression plasmid and specific siRNA against GUCD1 were obtained from RiboBio (Guangzhou, China). The corresponding vectors were transfected into HCC cells by Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) according to the manufacturers' proposals. At 48 h after transfection, the cells were harvested for further assay.
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate total RNA from HCC tissues and cell lines in line with the manufacturers' protocol. cDNA was synthesized from (2 µg) total RNA using PrimeScript™ RT Reagent kits (Takara Biotechnology Co., Ltd., Dalian, China). Then, cDNA was amplified with the SYBR Green Master Mix kit (Takara, Otsu, Japan) on the ABI 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Gene expressions were evaluated by the 2−ΔΔCT method. U6 and GAPDH were utilized as internal controls for miR-370 and GUCD1, as well as EMT-related genes, respectively. The sequences of the primers are described in Supplementary Table 1 (only online).
Total protein was isolated from cultured cells and lysed using RIPA lysis buffer (Thermo Fisher Scientific) containing protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Then, the protein concentration was quantified with a BCA protein assay kit (Beyotime Institute of Biotechnology, Shanghai, China). Subsequently, the proteins were subjected to separation by 10% SDS-PAGE and transferred onto PVDF membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% skim milk in TBST for 2 h at room temperature and incubated with specific primary antibodies overnight at 4℃. Afterwards, membranes were exposed to horse-radish peroxidase-linked secondary goat anti-rabbit antibody (1:2000, ab150077, Abcam, Cambridge, MA, USA) at room temperature for 1 h. The protein bands were visualized and detected with the enhanced chemiluminescence system (Pierce, Dallas, TX, USA). The primary antibodies were as follows: a rabbit antibody against GUCD1 (1:1000, sc-86315, Santa Cruz Biotechnology, Dallas, TX, USA), a rabbit antibody against E-cadherin (1:1000, ab40772, Abcam), a rabbit antibody against Vimentin (1:1000, ab92547, Abcam) and a rabbit antibody against GAPDH (1:2000, ab9485, Abcam). GAPDH was used as internal reference.
Transwell assay was conducted to detect the invasion and migration capacities of HCC cells. Briefly, Matrigel (BD Biosciences, San Jose, CA, USA) coated or uncoated transwell inserts (8.0 µm pore size, Corning Incorporated, Corning, NY, USA) were used to evaluate cell invasion or migration abilities. The following steps were the same for invasion and migration assays: Following 48 h of transfection, HCC cells were seeded in the upper chambers of the inserts in serum-free medium. At the same time, the bottom chamber was added with medium containing 10% FBS as chemoattractant. Being incubated for 48 h at 37℃ with 5% CO2, cells that remained on the surface of the top chambers were removed using cotton swabs, while cells attached to bottom chamber were fixed (10% methanol, 37℃, 15 min) and stained (0.1% crystal violet, 37℃, 10 min) for the detection under an inverted microscope (Olympus, Tokyo, Japan) from five randomly selected visual fields.
Wild-type or mutated GUCD13′-UTR sequences containing target sites of miR-370 were incorporated into the pGL3 vector (Promega, Madison, WI, USA) to obtain the wild-type GUCD1-3′UTR or mutant GUCD1-3′UTR, respectively. HCC cells were cotransfected with miR-370 mimics and luciferase reporter vectors of the wild- or mutant-type 3′-UTR of GUCD1 by Lipofectamine™ 2000 (Invitrogen). Subsequently, the Dual Luciferase Reporter Assay kit (Promega) was used to detect the relative luciferase activities at 48 hours after the transfection.
Statistical analysis was conducted with SPSS statistical software, version 17.0 (SPSS Inc., Chicago, IL, USA). Student's t test or ANOVA and Scheffe post-hoc test was used for comparison between two groups and multiple groups. p<0.05 was considered statistically significant difference. Correlation between mRNA and miRNA were estimated using the Spearman's correlation method.
We examined the expression levels of miR-370 and GUCD1 in HCC tissues and cells. Quantitative real-time PCR (qRT-PCR) demonstrated a dramatic decline of miR-370 expression in HCC tissues in comparison to normal tissues (Fig. 1A). Additionally, miR-370 expression in five HCC cells was notably downregulated, compared to normal liver cells (Fig. 1B). We investigated GUCD1 expression in the five HCC cell lines and found that GUCD1 was significantly upregulated when compared to the normal liver cells (Fig. 1C). Furthermore, to clearly demonstrate a correlation between GUCD1 and miR-370 in HCC, Spearman's correlation analysis was performed. As expected, the results demonstrated a negative correlation between GUCD1 and miR-370 expression in HCC (Fig. 1D).
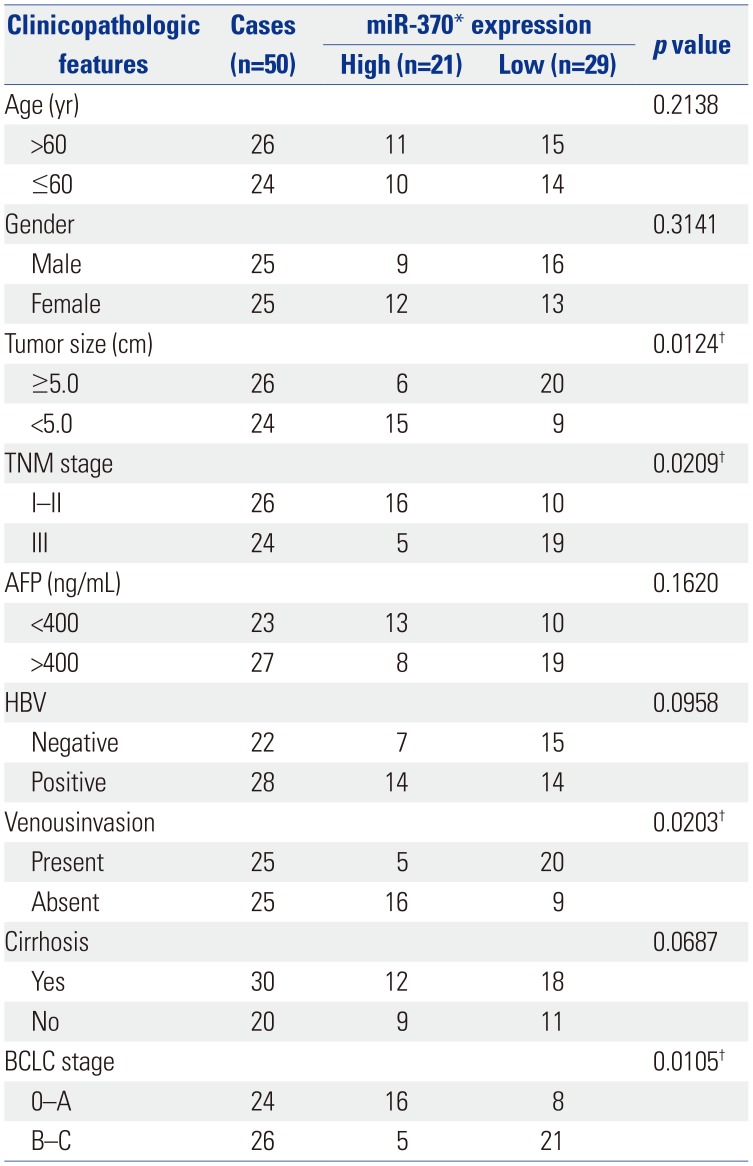
We next investigated the clinical significance of miR-370 to confirm its biological functions in HCC. Different subgroups of patients were plotted according to their miR-370 expressions; the mean level of miR-370 expressions was defined as the cutoff. As described in Table 1, the data indicated that decreased miR-370 expression was markedly associated with malignant HCC clinicopathologic characteristics. Moreover, Kaplan-Meier survival curves were used to analyze the survival rates of HCC patients. The HCC patients with low miR-370 expressions presented a notable poorer prognosis, compared to those with high miR-370 expression (Fig. 2A and B). Meanwhile, HCC patients highly expressing GUCD1 had shorter survival rates than their counterparts (Fig. 2C and D). In summary, our results revealed that GUCD1 and miR-370 maybe promising and potential biomarkers with which to predict the prognosis of HCC.
To explore the influence of miR-370 on HCC metastasis, miR-370 mimics or inhibitor was transfected into HCCLM3 and Hep3B cells, which had relatively lower and higher endogenous miR-370 expressions. qRT-PCR was performed to determine the transfection efficiencies and indicated that miR-370 expression in HCCLM3 cells was effectively upregulated while that in Hep3B cells was remarkably suppressed (Fig. 3A and B). Transwell assays were then carried out to verify that miR-370 overexpression could dramatically inhibit HCCLM3 cell invasion and that miR-370 knockdown markedly promoted Hep3B cell invasion (Fig. 3C). Transwell assays also indicated that miR-370 overexpression dramatically suppressed HCCLM3 cell migration, whereas miR-370 inhibition facilitated Hep3B cell migration (Fig. 3D).
EMT exerts significant functions in tumor invasion and migration. During the EMT process, mesenchymal markers, such as N-cadherin and vimentin, are upregulated, whereas epithelial markers, such as E-cadherin, are downregulated. In the present research, qRT-PCR and Western blot were carried out to investigate whether miR-370 regulates EMT markers. The results showed that expression of the EMT-related marker E-cadherin was upregulated, while N-cadherin and vimentin were downregulated (Fig. 4A and B) in HCCLM3 cells transfected with miR-370 mimics. Meanwhile, compared to the NC group, expression of E-cadherin was downregulated, while mRNA and protein expression of N-cadherin and vimentin was upregulated after transfecting miR-370 inhibitor into Hep3B cells (Fig. 4C and D). Thus, the data showed that miR-370 inhibited EMT of HCC cells.
To further elaborate on the mechanism underlying the suppressive function of miR-370 in HCC metastasis, TargetScan, miRanda, and PicTar were applied to search potential targets for miR-370, and the data showed that GUCD1 was one candidate gene that had complementary binding sites for miR-370 (Fig. 5A). Then, luciferase assays were performed to verify the association. Results indicated that miR-370 overexpression significantly inhibited the luciferase activity of wild-type GUCD1 3′-UTR, whereas it had no influence on the luciferase activity of mutant GUCD1 3′-UTR in HCC cells (Fig. 5B). Furthermore, we determined the regulatory functions of miR-370 in regulating GUCD1 expression in HCC cells by performing qRT-PCR and Western blots. The data indicated that miR-370 overexpression prominently decreased GUCD1 expression in HCCLM3 cells (Fig. 5C). Additionally, miR-370 inhibition remarkably increased GUCD1 expression in Hep3B cells (Fig. 5D). In short, these results demonstrated that GUCD1 was a direct target of miR-370 in HCC cells.
First, to confirm whether GUCD1 is a functional regulator of miR-370, GUCD1 overexpression plasmids were transfected into miR-370 overexpressed HCCLM3 cells. qRT-PCR and Western blots were then carried out to examine the transfection efficiencies (Fig. 6A). Subsequently, transwell assay was conducted, and the results demonstrated that GUCD1 restoration could dramatically abrogate the suppressive effects of miR-370 on HCCLM3 cell invasion and migration (Fig. 6B). Similarly, GUCD1 inhibition in miR-370-suppressed Hep3B cells could reverse the facilitating functions in Hep3B cell invasion and migration induced by miR-370 inhibitor (Fig. 6C and D).
Furthermore, we investigated whether alteration of GUCD1 expression could mediate the functions of miR-370 in HCC EMT. The results demonstrated that expression of the EMT-related markers N-cadherin and vimentin were upregulated and that E-cadherin expression was decreased (Fig. 7A and B) in miR-370 overexpressed HCCLM3 cells transfected with GUCD1 overexpression plasmids. Meanwhile, compared to the NC group, expression of E-cadherin was upregulated, while the expression of N-cadherin and vimentin was downregulated, after inhibiting GUCD1 expression in miR-370 suppressed Hep3B cells (Fig. 7C and D). Thus, these indicated that GUCD1 was a functional mediator of miR-370 in EMT of HCC cells.
HCC is a leading cause of tumor-associated deaths globally. Since it is hard to observe the clinical symptoms of HCC in the early stage, the prognosis for HCC, especially the advanced stage of HCC, is poor.22 Therefore, it is essential to explore new targets for HCC diagnosis and treatment. miRNAs have been confirmed to be closely related to HCC progression. For instance, miR-487a could promote HCC cell proliferation and metastasis23; miR-708 was found to suppress HCC by mediating SMAD324; and miR-135a was reported to promote cell invasion and migration in HCC via regulating forkhead box O1.25 All these evidence revealed that miRNA functions as crucial regulators in HCC.
Previous studies showed that miR-370 could regulate various human tumors: For example, miR-370 was found to regulate gastric cancer cell migration and proliferation by regulating EGFR.26 Studies by Han, et al.27 showed that alpinumisoflavone induced esophageal squamous cell carcinoma apoptosis via regulating miR-370/PIM1 signaling. Li, et al.28 found that miR-370 inhibited the Japanese encephalitis virus replication in glioblastoma. In the current study, we explored the functions of miR-370 in HCC and the results indicated that miR-370 was downregulated in HCC cells. Moreover, we further determined that miR-370 overexpression could suppress the activities of HCC cell lines by targeting GUCD1, including invasion, migration, and EMT. EMT is a key process that drives cancer metastasis, and the loss of E-cadherin and increase in N-cadherin and vimentin expression are considered to be the most important molecular markers of EMT. Recent studies have revealed that miRNAs act as crucial modulators of EMT through the regulation of relevant molecules. In our study, we demonstrated that miR-370 overexpression dramatically downregulated N-cadherin and vimentin expression and increased E-cadherin expression. In addition, reduced miR-370 expression was markedly related to poor prognosis and malignant clinicopathologic parameters of HCC patients. In short, the findings of this research demonstrated that miR-370 played important roles in HCC development.
The EMT process involves the activation of multiple signaling pathways, which are often interconnected and interacted with each other. The changes in various factors in the tumor microenvironment often promote tumor occurrence and progression. With the deepening of research on EMT-related signaling pathways and various related factors, the impact of EMT on HCC invasion and metastasis has been more widely recognized among researchers. However, there are currently no comprehensively recognized, sensitive, and specific EMT-related molecular markers. Liver biopsy or postoperative liver tissue specimen pathology can only evaluate EMT-related molecules in specific period of liver tissues, while the timing of EMT and the dynamic observation of EMT process are still difficult problems in EMT research. In addition, there is limited research on treatment of HCC based on the EMT mechanism. For instance, treatment regimens that block the TGF-β pathway and the Wnt/β-catenin pathway have certain effects on HCC.2930 These studies provide evidence for the dynamic observation of EMT in the progression and treatment of HCC, and provide a theoretical basis and new ideas for exploring new therapeutic targets of HCC.
Next, we explored the underlying mechanisms involved in the regulation of HCC by miR-370. GUCD1 is an important tumor-related modulator, and a previous study suggested that GUCD1 has an important influence on tumor development, including HCC.31 Here, the current study indicates that GUCD1 expression is inversely associated with miR-370 expression in HCC. We also found that inhibition of miR-370 significantly increases GUCD1 expression, while miR-370 overexpression remarkably decreases the expression of GUCD1 in HCC cells, indicating that GUCD1 is under the regulation of miR-370. In addition, our study also showed that GUCD1 is a target of miR-370 in HCC and modulates the repressive functions of miR-370 in HCC cell metastasis and EMT. Altering GUCD1 expression significantly reversed the functions of miR-370 in HCC cell invasion, migration, and EMT. Taken together, our data suggested that the miR-370/GUCD1 axis plays important roles in regulating HCC metastasis and EMT.
In conclusion, miR-370 is notably downregulated in HCC and its reduced expression is remarkably correlated with poor prognosis and malignant clinical parameters of HCC. Moreover, miR-370 overexpression dramatically suppresses HCC cell metastasis and EMT progression, whereas miR-370 inhibition markedly promotes them. Importantly, GUCD1 was identified as a target of miR-370. Moreover, GUCD1 restoration appears to abolish the functions of miR-370 in cell metastasis and EMT progression. In brief, miR-370 may function as a prognostic biomarker for HCC therapies.
Notes
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108. PMID: 25651787.

2. Kuper H, Ye W, Broomé U, Romelsjö A, Mucci LA, Ekbom A, et al. The risk of liver and bile duct cancer in patients with chronic viral hepatitis, alcoholism, or cirrhosis. Hepatology. 2001; 34(4 Pt 1):714–718. PMID: 11584367.

3. Wörns MA, Galle PR. HCC therapies--lessons learned. Nat Rev Gastroenterol Hepatol. 2014; 11:447–452. PMID: 24492278.

4. Jin Y, Li Q, Qiu J, Zhao X, Zheng C, Lv S, et al. Downregulation of paraoxonase 3 contributes to aggressive human hepatocellular carcinoma progression and associates with poor prognosis. Tumour Biol. 2016; 37:14193–14203. PMID: 27553024.

5. Wu X, Chen H, Gao Q, Bai J, Wang X, Zhou J, et al. Downregulation of JWA promotes tumor invasion and predicts poor prognosis in human hepatocellular carcinoma. Mol Carcinog. 2014; 53:325–336. PMID: 23169062.

6. Reichl P, Mikulits W. Accuracy of novel diagnostic biomarkers for hepatocellular carcinoma: an update for clinicians (review). Oncol Rep. 2016; 36:613–625. PMID: 27278244.

7. Lyons JG, Lobo E, Martorana AM, Myerscough MR. Clonal diversity in carcinomas: its implications for tumour progression and the contribution made to it by epithelial-mesenchymal transitions. Clin Exp Metastasis. 2008; 25:665–677. PMID: 18071912.

8. Sung WJ, Kim H, Park KK. The biological role of epithelial-mesenchymal transition in lung cancer (review). Oncol Rep. 2016; 36:1199–1206. PMID: 27460444.

9. Xue Y, Xu W, Zhao W, Wang W, Zhang D, Wu P. miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed Pharmacother. 2017; 86:426–433. PMID: 28012397.

10. Migita T, Ueda A, Ohishi T, Hatano M, Seimiya H, Horiguchi SI, et al. Epithelial-mesenchymal transition promotes SOX2 and NANOG expression in bladder cancer. Lab Invest. 2017; 97:567–576.

11. Gandellini P, Giovannetti E, Nicassio F. MicroRNAs in cancer management: big challenges for small molecules. Biomed Res Int. 2015; 2015:982156. PMID: 25879042.

12. Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012; 13:e249–e258. PMID: 22652233.

13. Garzon R, Marcucci G. Potential of microRNAs for cancer diagnostics, prognostication and therapy. Curr Opin Oncol. 2012; 24:655–659. PMID: 23079782.

14. Hu C, Lv L, Peng J, Liu D, Wang X, Zhou Y, et al. MicroRNA-375 suppresses esophageal cancer cell growth and invasion by repressing metadherin expression. Oncol Lett. 2017; 13:4769–4775. PMID: 28599478.

15. Guan H, Li W, Li Y, Wang J, Li Y, Tang Y, et al. MicroRNA-93 promotes proliferation and metastasis of gastric cancer via targeting TIMP2. PLoS One. 2017; 12:e0189490. PMID: 29220395.

16. Gao H, Cong X, Zhou J, Guan M. MicroRNA-222 influences migration and invasion through MIA3 in colorectal cancer. Cancer Cell Int. 2017; 17:78. PMID: 28855850.

17. Pan XP, Huang LH, Wang X. MiR-370 functions as prognostic marker in patients with hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2017; 21:3581–3585. PMID: 28925487.
18. Pan XP, Wang HX, Tong DM, Li Y, Huang LH, Wang C. miRNA-370 acts as a tumor suppressor via the downregulation of PIM1 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2017; 21:1254–1263. PMID: 28387905.
19. Bellet MM, Piobbico D, Bartoli D, Castelli M, Pieroni S, Brunacci C, et al. NEDD4 controls the expression of GUCD1, a protein upregulated in proliferating liver cells. Cell Cycle. 2014; 13:1902–1911. PMID: 24743017.

20. Della Fazia MA, Servillo G, Sassone-Corsi P. Cyclic AMP signalling and cellular proliferation: regulation of CREB and CREM. FEBS Lett. 1997; 410:22–24. PMID: 9247115.

21. Sands WA, Palmer TM. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008; 20:460–466. PMID: 17993258.

22. Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL, Hsieh TH, et al. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget. 2015; 6:23342–23357. PMID: 26160837.

23. Chang RM, Xiao S, Lei X, Yang H, Fang F, Yang LY. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. 2017; 23:2593–2604. PMID: 27827315.

24. Li Q, Li S, Wu Y, Gao F. miRNA-708 functions as a tumour suppressor in hepatocellular carcinoma by targeting SMAD3. Oncol Lett. 2017; 14:2552–2558. PMID: 28789462.

25. Zeng YB, Liang XH, Zhang GX, Jiang N, Zhang T, Huang JY, et al. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int. 2016; 16:63. PMID: 27486383.

26. Ning T, Zhang H, Wang X, Li S, Zhang L, Deng T, et al. miR-370 regulates cell proliferation and migration by targeting EGFR in gastric cancer. Oncol Rep. 2017; 38:384–392. PMID: 28534999.

27. Han Y, Yang X, Zhao N, Peng J, Gao H, Qiu X. Alpinumisoflavone induces apoptosis in esophageal squamous cell carcinoma by modulating miR-370/PIM1 signaling. Am J Cancer Res. 2016; 6:2755–2771. PMID: 28042498.
28. Li W, Cheng P, Nie S, Cui W. miR-370 mimic inhibits replication of Japanese encephalitis virus in glioblastoma cells. Neuropsychiatr Dis Treat. 2016; 12:2411–2417. PMID: 27703358.
29. Giannelli G, Villa E, Lahn M. Transforming growth factor-β as a therapeutic target in hepatocellular carcinoma. Cancer Res. 2014; 74:1890–1894. PMID: 24638984.

30. Huynh TT, Rao YK, Lee WH, Chen HA, Le TD, Tzeng DT, et al. Destruxin B inhibits hepatocellular carcinoma cell growth through modulation of the Wnt/β-catenin signaling pathway and epithelial-mesenchymal transition. Toxicol In Vitro. 2014; 28:552–561. PMID: 24434019.

31. Calvisi DF. Liver proliferation: the GUCD1/NEDD4-1 connection. Cell Cycle. 2014; 13:2022–2023. PMID: 24915571.

Fig. 1
miR-370 was downregulated, and GUCD1 was upregulated in HCC. (A and B) miR-370 expression in HCC tissues and cells was measured using qRT-PCR. (C) GUCD1 expression in HCC and normal liver cells was detected by qRT-PCR. (D) Spearman's correlation analysis between the miR-370 expressions and GUCD1 mRNA expressions in HCC tissues.*p<0.05, †p<0.01. GUCD1, guanylylcyclase domain containing 1; HCC, hepatocellular carcinoma.
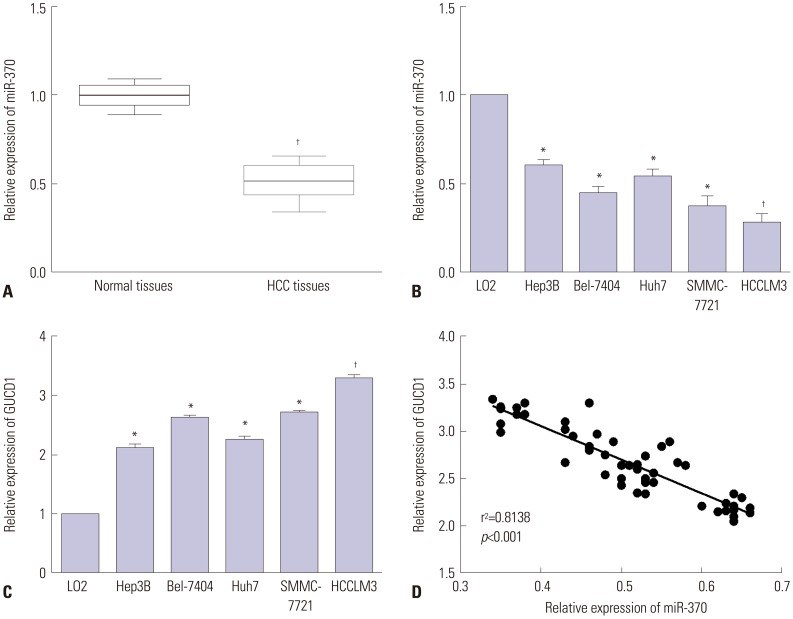
Fig. 2
Prognostic value of miR-370 and GUCD1 for HCC patients was analyzed by Kaplan-Meier analysis. (A) OS and (B) DFS of HCC patients with low and high miR-370 expression. (C) OS and (D) DFS of HCC patients with low and high GUCD1 expression. HCC, hepatocellular carcinoma; GUCD1, guanylylcyclase domain containing 1; OS, overall survival; DFS, disease-free survival.
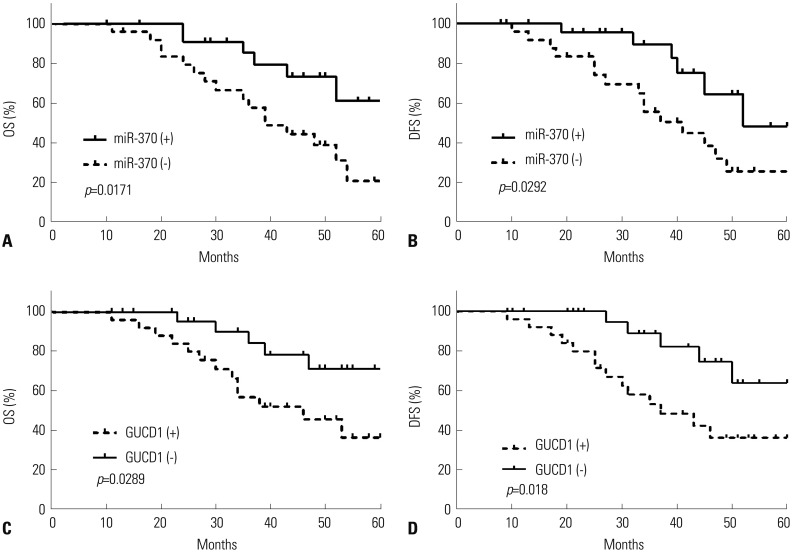
Fig. 3
miR-370 overexpression suppressed cell migration and invasion in HCC cells. (A) miR-370 expression in miR-370 overexpressed HCCLM3 cells were detected by qRT-PCR. (B) The expression levels of miR-370 in miR-370 suppressed Hep3B cells were examined using qRT-PCR. (C) Cell invasion and (D) migration abilities of miR-370 overexpressed or suppressed HCC cells were determined by transwell assays. *p<0.05, †p<0.01. HCC, hepatocellular carcinoma.
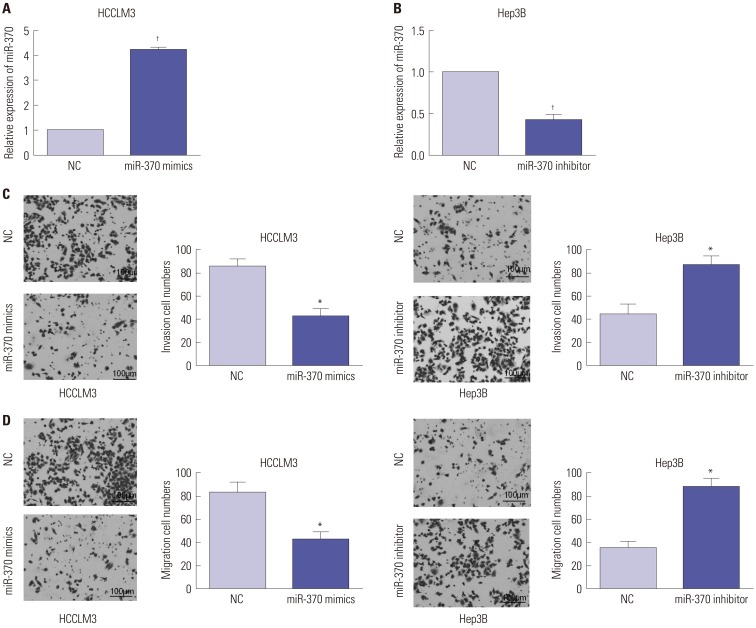
Fig. 4
miR-370 restoration suppressed EMT progression in HCC cells. (A) Western blot analysis and (B) qRT-PCR results of miR-370 overexpression in EMT of HCCLM3 cells. (C) Western blot and (D) qRT-PCR results indicated that inhibition of miR-370 in Hep3B cells downregulated E-cadherin expression and enhanced N-cadherin and Vimentin expression. *p<0.05, †p<0.01. EMT, epithelial-mesenchymal transition; HCC, hepatocellular carcinoma.
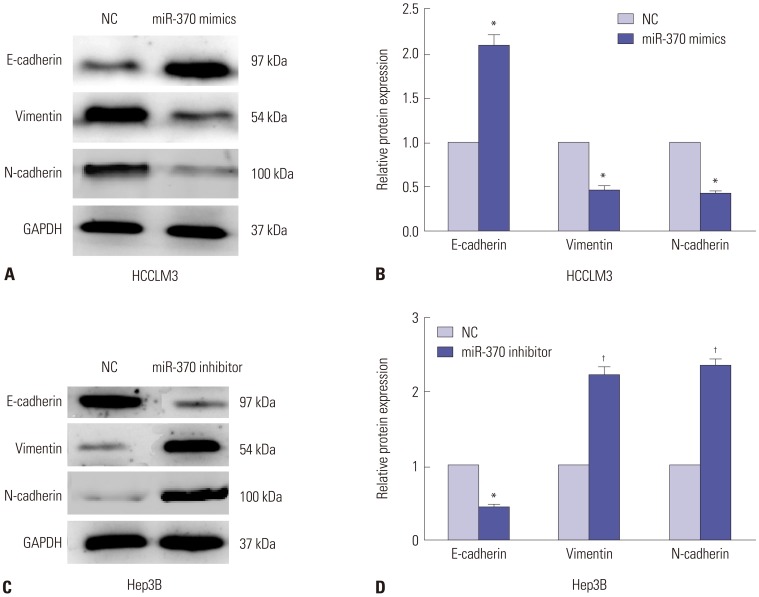
Fig. 5
GUCD1 was a direct target of miR-370 in HCC. (A) The putative binding sites of miR-370 in the GUCD1 3′-UTR. (B) Luciferase activity was detected by luciferase reporter gene assays in HCC cells cotransfected with wild-type or mutational GUCD1 3′UTR and miR-370 mimics, respectively. (C and D) GUCD1 expression in HCC cells transfected with miR-370 mimics or inhibitor were examined by Western blot (left) and qRT-PCR (right). *p<0.05. GUCD1, guanylylcyclase domain containing 1; HCC, hepatocellular carcinoma.

Fig. 6
Alteration of GUCD1 expression partially reversed the miR-370-mediated effect on HCC cell migration and invasion. (A) Western blot (up) and qRT-PCR (down) analysis of GUCD1 expression in miR-370-overexpressed HCCLM3 cells cotransfected with GUCD1 overexpression plasmid. (B) Transwell assays were conducted to examine cell migration and invasion abilities of miR-370-overexpressed HCCLM3 cells cotransfected with GUCD1 overexpression plasmid. (C) GUCD1 expression in miR-370-suppressed Hep3B cells cotransfected with GUCD1 siRNA was measured by Western blot (up) and qRT-PCR (down) analysis. (D) Transwell assays were performed to measure cell invasion and migration abilities of miR-370-suppressed Hep3B cells cotransfected with GUCD1 siRNA. *p<0.05. GUCD1, guanylylcyclase domain containing 1; HCC, hepatocellular carcinoma.

Fig. 7
Altering GUCD1 expression reversed the effect of miR-370 on cell EMT process in HCC. (A) Western blot and (B) qRT-PCR results showed that GUCD1overexpression in miR-370-overexpressed HCCLM3 cells decreased E-cadherin expression and increased N-cadherin and Vimentin expression. (C)Western blot and (D) qRT-PCR results showed that GUCD1 knockdown in miR-370-suppressed Hep3B cells upregulated E-cadherin and downregulated N-cadherin and Vimentin significantly. *p<0.05. GUCD1, guanylylcyclase domain containing 1; EMT, epithelial-mesenchymal transition; HCC, hepatocellular carcinoma.
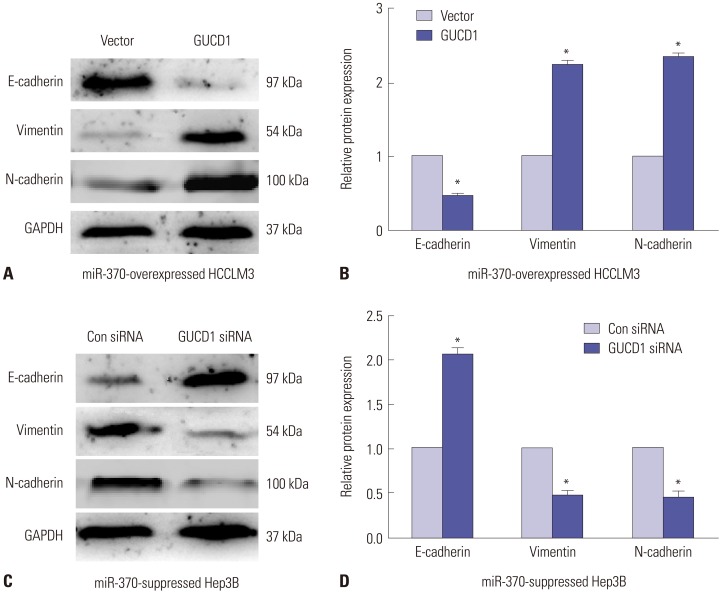
Table 1
Correlation of miR-370 Expression with the Clinicopathologic Characteristics of the Hepatocellular Carcinoma Patients




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download