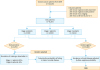
2. Statistical methods
To precisely estimate the probability of false-negative findings of the LN status due to missing a nodal disease, the data were simulated to reach the real situation to the greatest extent. Then, a β-binomial distribution was fitted to the simulated data to calculate this false-negative probability [
7]. Using the results derived from this modeling, NSS was finally calculated with 2 further steps:
1) Step 1: simulate the data of missing a detectable LN based on nodal-positive patients
To reach the real status of LNs metastasis more precisely, we performed the data simulation of missing a detectable LN.
A nodal positive patient indicated by the SEER database is a patient who has at least 1 pathologically metastatic lymph nodal. To precisely assess the probability of false negative findings—all of LNs examined were negative for a factually nodal positive patient, we established a dataset of false-negative patients containing 20% of pT1-2 patients and 40% of pT3 patients, respectively. Using a part of nodal-positive patients randomly selected from the dataset, we performed a data simulation as missing a LN that was randomly assigned for each patient. We assumed that n was the number of total LNs examined and i was the number of positive LNs examined for each patient; given n LNs examined, the patient was simulated as having 1 LN missed and therefore only n-1 LNs were examined. In the process of simulation, positive LNs for each patient were numbered from 1 to i and the negative LNs were numbered i+1 to n. Then, we generated a random positive integer termed rx from 1 to n, which was distributed uniformly. If rx≤i, the corresponding patient was simulated to have missed a detectable positive LN; otherwise in the condition of rx>i, the corresponding patient was simulated to have missed a detectable negative LN. Through the process of simulation, the dataset was assumed to contain false-negative patients. Then we construct a β-binomial model to assess the probability of false-negative findings due to missing a nodal disease based on the simulated dataset.
2) Step 2: compute the probability of failing to detect a nodal disease using a β-binomial distribution
In the following β-binomial model, we assumed that n was the number of total LNs examined from a patient, and i (i≥0) was the number of positive LNs examined based on the simulation data set. The β-binomial was listed below, where B (.) was a β function. The maximum likelihood method was used to estimate the parameters α and β.
In this model, k=n-i, represented the number of negative LNs for a patient. Data on jth patient was expressed as nj, ij, and kj, respectively (1≤kj<nj, j=1, 2,……N).
P (FNk) was the probability of all LNs examined negative (n=k, i=0) for an actually a LN-positive patient due to missing a nodal disease. To compute the probability of failing to detect a nodal disease, we assumed n=k in the above model. To ensure the accuracy for the fitted results, two assumptions should be fulfilled before the use of the statistical model s: firstly, it was the false-negative assumption that the positive LN was missed due to limited LNs examined intraoperatively; secondly, there was no false-positive results for LN pathological examination postoperatively.
3) Step 3: calculate the prevalence of nodal disease adjusted by false-negative probability
With the false-negative probability calculated from the above-mentioned steps, we estimated the adjusted prevalence of nodal disease stratified by pT stage:
For a given k- the total number of LNs excision, #TPk and #FNk represented the number of true nodal-positive and false nodal-negative patients.
Prev (Tj) represents the adjusted prevalence of nodal disease stratified by the pT stage. TNk is the number of true-negative patients.
4) Step 4: calculate the NSSs
Finally, NSSk|Tj(T=1, 2, 3) was calculated by the above-listed formula. It has 2 meanings of population- and individualized-based: the proportion of true nodal-negative EOC patients in pN0 patients with different pT stage, and the adequacy of a nodal-negative classification for those patients, respectively.
All statistical analysis was performed by using SAS (version 9.4; SAS Institute, Cary, NC, USA) and R (version 3.2.3; R Foundation, Vienna, Austria) software.







 PDF
PDF Citation
Citation Print
Print









 XML Download
XML Download