1. Lainscak M, Dagres N, Filippatos GS, Anker SD, Kremastinos DT. Atrial fibrillation in chronic non-cardiac disease: where do we stand? Int J Cardiol. 2008; 128(3):311–315.


2. Go AS, Hylek EM, Phillips KA, Chang Y, Henault LE, Selby JV, et al. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the AnTicoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. JAMA. 2001; 285(18):2370–2375.

3. Benjamin EJ, Wolf PA, D'Agostino RB, Silbershatz H, Kannel WB, Levy D. Impact of atrial fibrillation on the risk of death: the Framingham heart study. Circulation. 1998; 98(10):946–952.

4. Wolf PA, Abbott RD, Kannel WB. Atrial fibrillation as an independent risk factor for stroke: the Framingham study. Stroke. 1991; 22(8):983–988.


5. January CT, Wann LS, Alpert JS, Calkins H, Cigarroa JE, Cleveland JC. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: executive summary. A report of the American College of Cardiology/American Heart Association task force on practice guidelines and the Heart Rhythm Society. J Am Coll Cardiol. 2014; 64(21):2246–2280.
7. European Heart Rhythm Association. European Association for Cardio-Thoracic Surgery. Camm AJ, Kirchhof P, Lip GY, Schotten U, et al. Guidelines for the management of atrial fibrillation: the task force for the management of atrial fibrillation of the European Society of Cardiology (ESC). Eur Heart J. 2010; 31(19):2369–2429.
8. Thong T, McNames J, Aboy M, Goldstein B. Prediction of paroxysmal atrial fibrillation by analysis of atrial premature complexes. IEEE Trans Biomed Eng. 2004; 51(4):561–569.


9. Kirchhof P, Benussi S, Kotecha D, Ahlsson A, Atar D, Casadei B, et al. 2016 ESC guidelines for the management of atrial fibrillation developed in collaboration with EACTS. Eur Heart J. 2016; 37(38):2893–2962.


10. Aytemir K, Aksoyek S, Yildirir A, Ozer N, Oto A. Prediction of atrial fibrillation recurrence after cardioversion by P wave signal-averaged electrocardiography. Int J Cardiol. 1999; 70(1):15–21.


11. Clavier L, Boucher JM, Lepage R, Blanc JJ, Cornily JC. Automatic P-wave analysis of patients prone to atrial fibrillation. Med Biol Eng Comput. 2002; 40(1):63–71.


12. Blanche C, Tran N, Rigamonti F, Burri H, Zimmermann M. Value of P-wave signal averaging to predict atrial fibrillation recurrences after pulmonary vein isolation. Europace. 2013; 15(2):198–204.


13. Tateno K, Glass L. Automatic detection of atrial fibrillation using the coefficient of variation and density histograms of RR and deltaRR intervals. Med Biol Eng Comput. 2001; 39(6):664–671.

14. Sarkar S, Ritscher D, Mehra R. A detector for a chronic implantable atrial tachyarrhythmia monitor. IEEE Trans Biomed Eng. 2008; 55(3):1219–1224.


15. Dash S, Chon KH, Lu S, Raeder EA. Automatic real time detection of atrial fibrillation. Ann Biomed Eng. 2009; 37(9):1701–1709.

16. Lee J, Nam Y, McManus DD, Chon KH. Time-varying coherence function for atrial fibrillation detection. IEEE Trans Biomed Eng. 2013; 60(10):2783–2793.

17. Babaeizadeh S, Gregg RE, Helfenbein ED, Lindauer JM, Zhou SH. Improvements in atrial fibrillation detection for real-time monitoring. J Electrocardiol. 2009; 42(6):522–526.


19. Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS. Deep learning for visual understanding: a review. Neurocomputing. 2016; 187:27–48.

20. Angermueller C, Pärnamaa T, Parts L, Stegle O. Deep learning for computational biology. Mol Syst Biol. 2016; 12(7):878.

21. Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, et al. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017; 21(1):4–21.

22. Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015; 61:85–117.


23. Urtnasan E, Park JU, Joo EY, Lee KJ. Automated detection of obstructive sleep apnea events from a single-lead electrocardiogram using a convolutional neural network. J Med Syst. 2018; 42(6):104.


24. Kiranyaz S, Ince T, Gabbouj M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans Biomed Eng. 2016; 63(3):664–675.


25. Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000; 101(23):E215–20.
26. Van Alsté JA, Schilder TS. Removal of base-line wander and power-line interference from the ECG by an efficient FIR filter with a reduced number of taps. IEEE Trans Biomed Eng. 1985; 32(12):1052–1060.

27. Singh BN, Tiwari AK. Optimal selection of wavelet basis function applied to ECG signal denoising. Digit Signal Process. 2006; 16(3):275–287.

29. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Updated 2015.
https://arxiv.org/abs/1502.03167.
30. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014; 15:1929–1958.
33. Mohebbi M, Ghassemian H. Prediction of paroxysmal atrial fibrillation based on non-linear analysis and spectrum and bispectrum features of the heart rate variability signal. Comput Methods Programs Biomed. 2012; 105(1):40–49.


34. Boon KH, Khalil-Hani M, Malarvili MB, Sia CW. Paroxysmal atrial fibrillation prediction method with shorter HRV sequences. Comput Methods Programs Biomed. 2016; 134:187–196.











 PDF
PDF Citation
Citation Print
Print



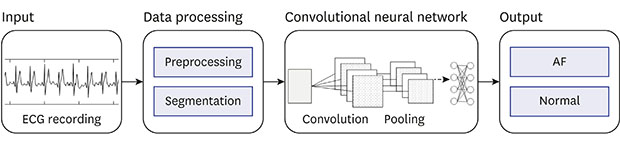





 XML Download
XML Download