Abstract
Pruritus (itching) is classically defined as an unpleasant cutaneous sensation that leads to scratching behavior. Although the scientific criteria of classification for pruritic diseases are not clear, it can be divided as acute or chronic by duration of symptoms. In this study, we investigated whether skin injury caused by chemical (contact hypersensitivity, CHS) or physical (skin-scratching stimulation, SSS) stimuli causes initial pruritus and analyzed gene expression profiles systemically to determine how changes in skin gene expression in the affected area are related to itching. In both CHS and SSS, we ranked the Gene Ontology Biological Process terms that are generally associated with changes. The factors associated with upregulation were keratinization, inflammatory response and neutrophil chemotaxis. The Kyoto Encyclopedia of Genes and Genomes pathway shows the difference of immune system, cell growth and death, signaling molecules and interactions, and signal transduction pathways. Il1a , Il1b and Il22 were upregulated in the CHS, and Tnf, Tnfrsf1b, Il1b, Il1r1 and Il6 were upregulated in the SSS. Trpc1 channel genes were observed in representative itching-related candidate genes. By comparing and analyzing RNA-sequencing data obtained from the skin tissue of each animal model in these characteristic stages, it is possible to find useful diagnostic markers for the treatment of itching, to diagnose itching causes and to apply customized treatment.
Pruritus can be caused by a variety of conditions, including skin disease. It can be divided as acute or chronic by time-dependent. Chronic pruritus is defined as pruritus lasting six or more weeks [1]. Acute itching is a sensation that occurs when the itch induction stimulus comes into contact with the skin and may be eliminated by pain in the periphery. That is why the pain is dominant and interacts with itching. However, chronic itching is a persistent sensation that occurs due to a variety of causes in which pain does not eliminate itching. Chronic itching itself causes scratching behavior and is an important starting point for the pathophysiology of skin damage. For these reasons, it is remains difficult to control itching despite extensive research and treatment efforts [2].
In variety of skin diseases, itch-scratch cycles are often continued, eliciting a skin barrier dysfunction. We noted two different mice models related to itching; contact hypersensitivity (CHS) and skin-scratching stimulation (SSS). A mouse model of CHS induced by 1-chloro-2,4-dinitrobenzene (DNCB) is a well-known allergic contact dermatitis model. DNCB is a kind of haptens, which is necessary to bind with proteins for immune response. When hapten is exposed into the skin, various chemical mediators, such as interleukin (IL)-1b and tumor necrosis factor (TNF), is released from keratinocytes. In contact allergy dermatitis, one of the most common symptoms is scratching behavior due to itching. Another mouse model called SSS was induced to become self-scratching models. Previous studies have shown that adding scratching stimuli on the dorsal skin of mouse after anesthesia can give rise to beginning to scratch themselves within a few minutes due to induction of substance P (SP) signaling. This SP/neurokinin 1 receptor (NK-1R) pathway is associated with pain and itch [345].
RNA-sequencing (RNA-seq) is one of the common techniques for the analysis of gene expression profiles and is recommended to accurate measurement of gene expression levels in skin lesions. More importantly, RNA-seq provides an unbiased view of the transcriptome [6].
Continuous stimulation initiates an inflammatory response as it dispatches the expression of numerous cytokines and controls neural signals by regulating the opening and closing of ion channels present at the sensory nerve endings. The transient receptor potential (Trp) pathway is a typical gene group that is involved in itching and pain sensation function [7].
In this study, we investigated whether skin injury caused by chemical/physical stimuli causes initial pruritus, and analyzed gene expression profiles systemically to determined how changes in skin gene expression in the itching area are related to itching.
Female CBA/J mice (8 weeks old, 20 ± 2 g; KOATECH, Pyeongtaek, Korea) were used. Breeding was carried out in an individual ventilation cage system in a general animal breeding room of Chung-Ang University. A ventilation mouse cage (192 × 372 × 131 mm) was used for a breeding box, and five animals were placed in each group. The litter was fed with Coarse Sani-Chips (ENVIGO, East Millstone, NJ, USA). The animals were fed freely (Purina Mouse Diets, Seongnam, Korea) and watered freely. The experiment started after the completion of the review of the Institutional Animal Care and Use Committee (IACUC) in Chung-Ang University (IACUC approval No. 2018-00082 and 2018-00083).
Chloroform-isoamyl alcohol (Cat. No. 25666) and olive oil (Cat. No. 1514) were purchased from Sigma-Aldrich (St. Louis, MO, USA), and acetone (Cat No.1.07021.2511), 2-propanol (Cat. No.1.09634.1011), and ethanol (Cat. No. 1.00983.1011) were purchased from Merck (Darmstadt, Germany). 3M sodium acetate (pH 5.2) (Cat. No. C-9008) was purchased from Bioneer (Daejeon, Korea). Alfaxan and Rompun injection were obtained from Jurox Pty Ltd. (Rutherford, Australia) and Bayer Korea (Seoul, Korea), respectively.
The CHS model was modified from a local lymph node assay (OECD Test Guideline 429) [3]. Body weight was measured the day before application and animals were randomly divided into 2 groups (5 mice per group). After anesthesia, the hair in dorsal skin was removed using a depilatory cream (Body Natur cream; Viokox S.A., Aldaia, Spain) and a clipper. The stimulant application start date was set to 0 day, and it was applied once a day for a predetermined time through 0 to 2 days of the test. The stimulants were prepared by dissolving DNCB in a mixture of acetone and olive oil (AOO; acetone:olive oil = 4:1), which served as a vehicle, in a final concentration of 1% (w/v). This stimulant (25 µl) was administrated to mice. To confirm the effect of vehicle, a vehicle control (VT) of AOO only, was also applied (Supplementary Fig. 1).
The SSS model was modified from a previous study [5]. Briefly, the body weight was measured the same day as the CHS model and the hair was removed using a depilatory cream (Body Natur cream) and a clipper after anesthesia. Skin irritation was performed once with an artificial brush using an artificial stimulation condition (scratch stimulation pressure, 100 gf/cm2; scratch stimulation frequency, 90 times/min; scratch stimulation duration, 10 min). It was observed whether the mice acted spontaneous scratching behavior within minutes after the SSS terminated. The non-treated control (NT) group was an SSS control group and only anesthesia and epilation were performed (Supplementary Fig. 1).
The animals were acclimated in a Plexiglas chamber (diameter, 95 cm; height, 175 cm) for 10 min, followed by video observation for 10 min. One scratching was defined as the time where the hind legs were lifted to scratch the skin irritation site and then touched the ground or mouth again. VT and CHS performed a scratching behavior observation at constant time. SSS was recorded for 10 min when spontaneous scratching was observed after skin stimulation. If no scratching behavior was observed within 20 min after the end of the stimulation, the observation was terminated [8].
When the next day after giving the last stimulus, the animal was sacrificed. The dorsal skin tissues were cut to 1 × 2 cm and sent to Macrogen Inc. (Seoul, Korea) for RNA-seq. The extraction of total RNA from tissue by using the TRIzol® Reagent (15596026; Invitrogen, Carlsbad, CA, USA) was conducted according to the user manual. We check total RNA integrity using a 2200 TapeStation with an RNA integrity number value greater than or equal to 6.3. The experimental conditions were as follows; paired-end (type of read), 151 bp (read length), TruSeq Stranded mRNA LT Sample Prep Kit (library kit), TruSeq Stranded mRNA Sample Preparation Guide, Part # 15031047 Rev. E (library protocol), using an Illumina platform (type of sequence; San Diego, CA, USA). To assure the sequencing performance, we used RNA-SeQC [9] to assess the RNA-seq data quality for each sample and the raw RNA-seq data were filtered by removing adaptor sequences, contamination, and low-quality reads. For each sample, we quantified the expression values of all known mRNA transcripts in the Ensembl [10] mouse gene annotation (GRCm38) using Sailfish [11], with default settings. For each gene, the transcripts per million value was recorded. The RNA-seq data were used to prioritize the itching related genes.
The edgeR [12] algorithm was used to identify the differentially expressed genes from the RNA-seq data. The genes with a false discovery rate (FDR) < 5% and fold change (FC) > 2 were deemed differentially expressed. Based on Gene Ontology Biological Process (GOBP) definition [13], Fisher's exact test was applied to determine whether the proportion of differentially expressed genes in one given GOBP geneset is significantly enhanced.
The FAIME algorithm calculates geneset scores using the rank-weighted gene expression of individual samples, which converts each sample's transcriptomic profile to molecular mechanisms [14]. A higher pathway score suggests an overall increase in the expression of the genes within the given pathway. Student t-test was used to identify the differentially expressed KEGG pathways between VT and CHS samples and between NT and SSS. The pathways with adjusted p < 0.05 (by Benjamini-Hochberg procedure) were deemed differentially expressed.
The STRING database (ver. 10.5; https://string-db.org/) is one of the search tools for the interaction between genes/proteins. We predicted the PPI of Trp ion channels and cytokines, which were selected based on literature review and RNA-seq results, were analyzed. The PPI was limited to an interaction threshold of 0.4 (medium confidence) [15].
Body weight data were expressed as mean and standard deviation (mean ± standard deviation), and one-way ANOVA was used for comparison between the groups. Scratching was scored on an individual basis and results were subjected to a one-tailed test using a paired sample t-test. The data were analyzed using SPSS Statistics (version 20; IBM, Armonk, NY, USA). Other statistical methods according to each experiment are mentioned separately.
Total RNA was isolated from dorsal skin tissue of both models, and RNA-seq analysis was performed to observe the RNA change pattern observed in each model. At the specified significance level (FC > 2 and FDR < 0.05), 292 genes were found to be commonly upregulated in both the CHS and SSS groups, compared with the VT and NT samples, respectively. In both CHS and SSS, we ranked the GOBP terms that are significantly associated with the transcriptomic changes (Fig. 1A). The KEGG pathway showed the difference of immune system, signaling molecules and interaction (Fig. 1B). The upregulated genes corresponding to the GO and KEGG pathways were identified. Il1b was upregulated both CHS (log2FC = 3.6) and SSS (log2FC = 6.9). Il1a (log2FC = 2.0), Il22 (log2FC = 3.8) were upregulated in CHS and Tnfrsf1b (log2FC = 2.0), Il6 (log2FC = 9.1) and Il1r1 (log2FC = 2.0) were upregulated in SSS (Figs. 2A and 3).
Because the Trp ion channel is closely related to itching, we observed changes in the expression of all Trp ion channels (28 genes, Supplementary Table 1). The criteria for selection of the differentially expressed genes is a log2FC of greater than 1.2 or less than −1.2, and then an FDR of less than 0.05. Trpm3 was downregulated both CHS (log2FC = −1.5) and SSS (log2FC = −1.4). Trpc1 (log2FC = −1.5) was downregulated in CHS. However, Trpm1 (log2FC = 2.9), Trpm2 (log2FC = 2.3) and Mcoln2 (log2FC = 3.8) were upregulated and Trpv6 (log2FC = −1.6) was downregulated in SSS (Fig. 2B). These results could be seen that Trpc1 is CHS-specific and Trpv6, Trpm1, Trpm2 and Mcoln2 are SSS-specific Trp channels.
Network analysis was uploaded all the differentially expressed genes (13 genes, |log2FC| > 1.2 and FDR < 0.05) which identified knowledge and RNA-seq data. Using an interaction threshold of 0.4 (medium confidence), the network analysis shown a highly clustered network (clustering coefficient, 0.779) containing 26 edges (expected number of edges, 2; PPI enrichment p-value, 1.0 × 10−16). There were 108 GOBP and 36 KEGG pathways. The genes in each cluster were highly correlated. Between the gene cluster of inflammation and the gene cluster of itching related ion channels were less specific and less associated (Fig. 4).
The itching theory is generally accepted as an intensity theory resulting from the weak activation of the sensory fibers of the skin. In contrast to this hypothesis, the specificity theory explaining the presence of distinct neurons with separate pathways from itching and pain and the nociceptors capable of activating the central neurons responsive to itching has greater support. There is also a pattern theory that explains the selectivity theory, or that there is no special circuit for any particular sensation; that all senses are encoded in a specific activation pattern of sensory neurons, and when perceived, they cause a perceived sense of perception [16]. To date, it has been suggested that the nerve endings of the epidermis are involved in enhancing the senses because there is no specific receptor that distinguishes itching and pain nerves. The receptors distributed in the skin sense general stimuli such as tactile, nociceptive, angular, and temperature and are transmitted to the coarse nerve bundles in the subcutaneous tissue [17].
We confirmed, through molecular genetic techniques, the starting point of the sense of itching in CHS and SSS mice models, which were selected in order to block the cause of skin damage due to scratching behavior. Although these two models have common features of skin barrier damage and inflammatory response, the patterns of expression of several genes were different. The SSS model was first proposed by the Seiji Kawana team in Japan in 2007 [5]. This model has been reported to induce physiological skin stimulation in ICR mice increasing the nerve growth factor (NGF) signaling pathway and to mediating a portion of the SP signaling pathway through NK-1R. NGF is produced in various skin cells and increased in expression in response to IL-1, IL-2, and ultraviolet rays. In addition, NGF induces degranulation of mast cells and stimulates neuronal fiber germination and sensory nerve proliferation, thereby increasing the itch sensation in skin diseases. We confirmed whether the SSS model reproduced is similar to the previous research model in two ways. One was behavioral observation and the other was gene expression confirmation. Unlike the expectation, scratching behavior was not observed in NT and SSS (Fig. 5B) and expression levels of Tac1 and Tacr1 were similar in NT and SSS (data not shown). Both of CHS and SSS animal models gained in weight normally and showed no change in body weight by itching related stimuli (Fig. 5A). These results could be explained that it was a general reaction for the following reasons. When skin barriers are damaged in normal skin, cytokines and growth factors that are expressed early in inflammation release pro-inflammatory mediators, and neutrophils are recruited by these chemotactic factors [18]. Neutrophils are activated the inflammatory stage of wound healing. In the present study, it was assumed that an increase in signal transduction related to neutrophils would be observed, indicating the onset in the early stage of inflammation. This causes Il1a to initiate an inflammatory response. Il1b, an amplification factor for inflammation [19], then increases the pro-inflammatory response with Tnf, whereby Il6 and Tnf act as ideal markers for rapidly initiating inflammation [20]. The SSS model showed the same index as this normal inflammatory response. On the other hand, in the case of CHS, the expression of Il22 and its receptor, Il22ra1, increased. Il22 is mainly expressed by immune system cells, affects non-hematopoietic cells, and is overexpressed in patients with atopic dermatitis with acute and chronic atopic lesions. Il22 also enhances wound healing in the skin [21]. Decreased cosynergism of Tnf and Il22 showed defects in neutrophil recruitment in Il22 deficient mice (Figs. 2A and 3).
TRPs ion channels are nonselective cation channels that are activated by stimuli that cause pain. Trpv1 and Trpa1 were the focus of many studies in relation to itching [22]. Trpv1 is known to be a mediator of histamine-induced itching. Trpv1 was activated directly or causes itching via the PLA2/LO pathway. Trpa1 is involved in non-histaminergic itching with Tslp (thymic stromal lymphopoietin) [23]. Trpa1 activates neuronal excitability by Tslp or stimulates Orai1/NFAT signal by the release of Tslp. In this study, however, there was little or no change in expression of Trpv1 and Trpa1. This is interpreted as a result of the correlation between the temperature of the initial stimulus and Trp expression because the Trp ion pathway is thermosensitive or mechanosensitive [2]. It has been confirmed that the Trp ion channel is involved by examining the pre-itching generator and skin diseases (Figs. 2B and 3). We have noticed is Trpc1. The increase of Trpc1 is known to increase the epithelial cell proliferation. Trpc1 also is involved in the regulation of cytosolic Ca2+ and Na+ in astrocytes and the abolition of Trpc1 induces astrogliosis [24]. Recent studies suggest that astrogliosis [25] is associated with chronic itching. However, the relevance of astrogliosis-like to neurons in the skin needs to be studied. Trpm1, Trpm2, Trpm3, Trpv6 also detected association in this study. There are few studies reporting that these channels are associated with itching. One possibility is the association with the inflammatory response. In particular, Mcoln2 [26] is related to the innate immune response and affects the entire wound healing process (Fig. 4).
Although there is a clear limitation to the use of models with different stimulation durations and methods, the focus of this study was to identify the overall changes in skin genes at the time of initial itching with an aim at providing data for future studies and to aid in the treatment planning for itching.
In this study; 1) SSS quickly reached an inflammation reaction leading to the wound treatment phase instead of the itching step, which causes the scratching action due to the duplication of pain signals (Tnf and SP), resulting in the normal wound treatment result; 2) Results of the CHS model suggests that the initial inflammatory signal is reduced, the sequential wound healing process is delayed, and if this condition persists, it eventually progresses to the itching stage; 3) By comparing and analyzing the RNA-seq data obtained from the skin tissue of each animal model in these characteristic stages, it was possible to find useful diagnostic markers for the treatment of itching, to diagnose causes of itching, to apply customized treatment. We expect this data to be used as useful in the future of itching treatment.
Figures and Tables
 | Fig. 1Overview of RNA-sequencing based characterization.(A) The top 10 Gene Ontology Biological Process terms associated with the commonly upregulated genes. The p-value were computed by Fisher's exact test and corrected by Bonferroni procedure. The vertical dash line denotes the significance level of α = 0.05. (B) Heatmaps of genes differentially expressed between CHS and SSS. Each column in the heatmap denotes one mouse while each row denotes one Kyoto Encyclopedia of Genes and Genomes pathway. Red represents relatively increased gene expression while blue represents down-regulation. A t-test was used to prioritize the pathways differentially expressed between suitable control and case samples (adjusted p < 0.05). The pathway expression score was computed by the FAIME algorithm. CHS, contact hypersensitivity group; SSS, skin-scratching stimulation group; VT, vehicle control group; NT, non-treated control group.
|
 | Fig. 2Bubble chart representing the relationship of the gene expression between CHS and SSS.(A) Changes in the genes associated with inflammation. (B) Changes in all of the Trp channel genes. X-axis, log2FC between NT and SSS; Y-axis, log2FC between VT and CHS. The bubble size was the sum of absolute value of log2FC for representative of the genes attributed to expression levels. CHS, contact hypersensitivity group; SSS, skin-scratching stimulation group; FC, fold change; NT, non-treated control group; VT, vehicle control group.
|
 | Fig. 3Boxplot of candidate genes related pruritus.Y-axis represented the expression level by transcripts per million (TPM). Last two plots showed two genes with housekeeping profile. CHS, contact hypersensitivity group; SSS, skin-scratching stimulation group; NT, non-treated control group; VT, vehicle control group.
|
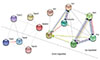 | Fig. 4Network analysis of candidate itching related genes.The differentially expressed genes (13 genes) which identified knowledge and RNA-sequencing data were uploaded. The nodes were genes and linear edges indicated molecular action for association in published data sets and line shape indicates the predicted mode of action.
|
 | Fig. 5In-vivo data.(A) Changes in the body weight. The body weight was increased generally and there was no difference between each group. The final body weight was measured after fasting the day before the sacrifice. (B) Changes in scratching behavior VT and CHS shown scratching behavior. There was difference compared with before and after stimulation, statistically, in CHS (*p < 0.05). Olive oil, materials of AOO, itself may cause contact allergy. Some differences may exist between NT and VT. CHS, contact hypersensitivity group; SSS, skin-scratching stimulation group; NT, non-treated control group; VT, vehicle control group; AOO, a mixture of acetone and olive oil.
|
ACKNOWLEDGEMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A1B03932890). This research was supported by the Chung-Ang University Research Grants in 2018.
Notes
Author contributions Y.W.K., E.A.K. and T.Z. designed the experiments and interpreted the data. N.K., T.C., H.L., S.C., G.B., Y.H., D.K., H.P., M.L., E.J., J.C., Y.S., S.K., and Y.W.K. performed experiments. D.L., H.M.B., and I.L. analyzed the data. Y.W.K. and T.Z. wrote the manuscript. H.W.B. and J.H.K. organized the study.
References
1. Ständer S, Weisshaar E, Mettang T, Szepietowski JC, Carstens E, Ikoma A, Bergasa NV, Gieler U, Misery L, Wallengren J, Darsow U, Streit M, Metze D, Luger TA, Greaves MW, Schmelz M, Yosipovitch G, Bernhard JD. Clinical classification of itch: a position paper of the International Forum for the Study of Itch. Acta Derm Venereol. 2007; 87:291–294.


2. Moore C, Gupta R, Jordt SE, Chen Y, Liedtke WB. Regulation of pain and itch by TRP channels. Neurosci Bull. 2018; 34:120–142.


3. Anderson SE, Siegel PD, Meade BJ. The LLNA: a brief review of recent advances and limitations. J Allergy (Cairo). 2011; 2011:424203.

4. Honda T, Egawa G, Grabbe S, Kabashima K. Update of immune events in the murine contact hypersensitivity model: toward the understanding of allergic contact dermatitis. J Invest Dermatol. 2013; 133:303–315.


5. Yamaoka J, Kawana S. A transient unresponsive state of self-scratching behaviour is induced in mice by skin-scratching stimulation. Exp Dermatol. 2007; 16:737–745.


6. Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA. 2017; 8:e1364.

7. Zhang X. Targeting TRP ion channels for itch relief. Naunyn Schmiedebergs Arch Pharmacol. 2015; 388:389–399.


8. Shim WS, Tak MH, Lee MH, Kim M, Kim M, Koo JY, Lee CH, Kim M, Oh U. TRPV1 mediates histamine-induced itching via the activation of phospholipase A2 and 12-lipoxygenase. J Neurosci. 2007; 27:2331–2337.
9. DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012; 28:1530–1532.



10. Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, et al. Ensembl 2015. Nucleic Acids Res. 2015; 43:D662–D669.

11. Patro R, Mount SM, Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol. 2014; 32:462–464.



12. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140.


13. Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004; 32:D258–D261.
14. Yang X, Regan K, Huang Y, Zhang Q, Li J, Seiwert TY, Cohen EE, Xing HR, Lussier YA. Single sample expression-anchored mechanisms predict survival in head and neck cancer. PLoS Comput Biol. 2012; 8:e1002350.

15. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–D452.

17. Twycross R, Greaves MW, Handwerker H, Jones EA, Libretto SE, Szepietowski JC, Zylicz Z. Itch: scratching more than the surface. QJM. 2003; 96:7–26.


18. Rittié L. Cellular mechanisms of skin repair in humans and other mammals. J Cell Commun Signal. 2016; 10:103–120.



19. Jensen LE. Targeting the IL-1 family members in skin inflammation. Curr Opin Investig Drugs. 2010; 11:1211–1220.


20. Wang XP, Schunck M, Kallen KJ, Neumann C, Trautwein C, Rose-John S, Proksch E. The interleukin-6 cytokine system regulates epidermal permeability barrier homeostasis. J Invest Dermatol. 2004; 123:124–131.


21. McGee HM, Schmidt BA, Booth CJ, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA, Horsley V. IL-22 promotes fibroblast-mediated wound repair in the skin. J Invest Dermatol. 2013; 133:1321–1329.

22. Feng J, Yang P, Mack MR, Dryn D, Luo J, Gong X, Liu S, Oetjen LK, Zholos AV, Mei Z, Yin S, Kim BS, Hu H. Sensory TRP channels contribute differentially to skin inflammation and persistent itch. Nat Commun. 2017; 8:980.



23. Wilson SR, Thé L, Batia LM, Beattie K, Katibah GE, McClain SP, Pellegrino M, Estandian DM, Bautista DM. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell. 2013; 155:285–295.



24. Belkacemi T, Niermann A, Hofmann L, Wissenbach U, Birnbaumer L, Leidinger P, Backes C, Meese E, Keller A, Bai X, Scheller A, Kirchhoff F, Philipp SE, Weissgerber P, Flockerzi V, Beck A. TRPC1- and TRPC3-dependent Ca2+ signaling in mouse cortical astrocytes affects injury-evoked astrogliosis in vivo. Glia. 2017; 65:1535–1549.


SUPPLEMENTARY MATERIALS
Supplementary data including one table and one figure can be found with this article online at http://pdf.medrang.co.kr/paper/pdf/Kjpp/Kjpp2019-23-02-07-s001.pdf.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download