Abstract
Canine MDR1 gene mutations produce translated P-glycoprotein, an active drug efflux transporter, resulting in dysfunction or over-expression. The 4-base deletion at exon 4 of MDR1 at nucleotide position 230 (nt230[del4]) in exon 4 makes P-glycoprotein lose function, leading to drug accumulation and toxicity. The G allele of the c.-6-180T>G variation in intron 1 of MDR1 (single nucleotide polymorphism [SNP] 180) causes P-glycoprotein over-expression, making epileptic dogs resistant to phenobarbital treatment. Both of these mutations are reported to be common in collies. This study develops a more efficient method to detect these two mutations simultaneously, and clarifies the genotype association with the side effects of chemotherapy. Genotype distribution in Taiwan was also investigated. An oligonucleotide microarray was successfully developed for the detection of both genotypes and was applied to clinical samples. No 4-base deletion mutant allele was detected in dogs in Taiwan. However, the G allele variation of SNP 180 was spread across all dog breeds, not only in collies. The chemotherapy adverse effect percentages of the SNP 180 T/T, T/G, and G/G genotypes were 16.7%, 6.3%, and 0%, respectively. This study describes an efficient way for MDR1 gene mutation detection, clarifying genotype distribution, and the association with chemotherapy.
Mammalian P-glycoprotein is an active drug efflux transporter located in the plasma membrane, and the MDR1 gene regulates the expression of P-glycoprotein. MDR1 knock-out mice showed a profound effect on the accumulation of a range of drugs. P-glycoprotein prevents the accumulation of some drugs, which is a prerequisite for their clinical use, and absence of the transporter can lead to severe toxicity [3]. P-glycoprotein is an integral part of the blood-brain barrier and is essential in keeping large, hydrophobic drugs out of the brain. Moreover, P-glycoprotein in the small and large intestines reduces intestinal uptake (oral drug availability) and mediates direct intestinal excretion of substrates. P-glycoproteins in liver and kidney mediate hepatobiliary and renal excretion of substrates, respectively [3]. In addition, the over-expression of P-glycoprotein in tumor cells can contribute to multidrug resistance [35].
A 4-base deletion at exon 4 of the MDR1 gene at nucleotide position 230 (nt230[del4]) results in a frame shift, generating several stop codons that prematurely terminate P-glycoprotein synthesis [813]. Dogs that are homozygous for that deletion mutation display ivermectin-sensitive [13] or moxidectin-sensitive [8] phenotypes. The MDR1 gene 4-base deletion is found mostly in collie breeds [16].
The single nucleotide polymorphism (SNP) 180 mutation of the MDR1 gene is at the G allele of the c.-6-180T>G variation in intron 1 and is significantly more frequent in epileptic Border Collies resistant to phenobarbital treatment, which is related to P-glycoprotein over-expression [1]. The G allele frequency has been reported to be extremely high in Border Collies [17].
Detection of the 4-base deletion genotype and SNP 180 genotype of the MDR1 gene is important in dogs. Many detection methods have been developed, including the use of mutagenetically separated polymerase chain reaction (PCR) [16], real-time PCR [11], and restriction fragment length polymorphism (RFLP) [19] for 4-base deletion detection, and sequencing [1] and RFLP [17] for SNP 180 detection. However, no integrated detection of both mutations has been reported. In this study, we developed a simple approach to detect these two MDR1 gene mutations simultaneously by using an oligonucleotide microarray system. Furthermore, the distributions of the 4-base deletion and SNP 180 genotypes are unreported in Taiwan, as are the correlations between the two genotypes and the relevant chemotherapy side effects. The purpose of this study was to clarify these issues. The developed microarray-based method can provide instant genotype identity messages efficiently, assisting in medication choice and dosage determination by clinicians and furthering the prevention of chemotherapy side effects.
The gene sequences corresponding to the 4-base deletion and the SNP 180 substitution in the canine MDR1 gene were identified by comparing the published primer pairs [1117] and the canine MDR1 gene sequences (GenBank accession Nos. NM_001003215.2 and NM_001003215.2) using DNASTAR lasergene (DNASTAR, USA), respectively. The primers and probes for the microarray tests were designed and are as shown in Tables 1 and 2, respectively. Sequences containing the 4-base deletion (MM genotype), the 4-base no-deletion wild-type (WW genotype), the SNP 180 substitution (G allele) and original (T allele) nucleotides encompassing the corresponding forward and reverse primer loci were synthesized artificially as the standards.
Singlet and multiplex PCR were performed using Fast-Run Taq Master Mix kits (Protech, Taiwan) containing pfu polymerase with a nucleotide proofreading function. The singlet and multiplex PCR conditions were the same except for the primer numbers and were carried out in a reaction volume of 50 µL containing 25 µL 2× Taq Master Mix, 150 ng/dL template DNA, and 2 µL of each primer (10 µM). The thermal profile for amplification was 94℃ for 3 min, 34× (94℃ for 45 sec, 58℃ for 30 sec, and 72℃ for 40 sec), and 70℃ for 5 min. The 4-base deletion genotype PCR products were separated in 5% agarose gel at 100 V for 8 h or 12% polyacrylamide gel electrophoresis (PAGE) gel at 50 V for 8 h. The SNP 180 genotype PCR products and the multiplex PCR products were separated in 2% agarose gel at 100 V for 40 min. The gels were stained with ethidium bromide and visualized under UV light.
A tail composed of 15 T bases was added to each 5′ end of the oligonucleotide probe, including that of the positive control probe (an oligonucleotide from capsid protein VP1 of human enterovirus 71 gene, 5′-ATGAAGCATGTCAGGGCTTGGATACCTCG-3′). Ten microliters of each probe were then spotted to each specific position on the microarray polymer substrate by using an automatic spotting machine and immobilized using a UV crosslinker (UV Stratalinker 1800; Strategene, USA) at 8,000 µJ. Hybridization reactions between the DNA templates and their corresponding probes were carried out using the Dr. Chip DIY Kit (Dr. Chip Biotech, Taiwan). The procedures followed the supplier's manual and are described briefly below. Multiplex PCR was performed using 5′-end biotin-labeled primers. The PCR product was denatured at 94℃ for 10 min and then cooled in an ice bath for 5 min. To the microarray chamber were added 200 µL of hybridization buffer (containing the 5′-end biotinylated oligonucleotide that was complementary to the positive control probe sequence) and 10 µL of denatured PCR product, followed by incubation at 51℃ with vibration for 1 h. The sample was washed three times with washing buffer. Strep-AP (Streptavidin conjugate alkaline phosphatase; Dr. Chip Biotech, Taiwan) was diluted 1,000 times by volume in blocking reagent, and, for the blocking reaction, 200 µL Strep-AP was added to the chamber at room temperature for 30 min. The chamber was washed three times with washing buffer. A colorimetric reaction was then induced by adding 4 µL of NBT/BCIP and 196 µL of detection buffer in the chamber, followed by developing in the dark at room temperature for 5 min, and washing twice with distilled water. The hybridization result was indicated by the developed pattern on the microarray, which was read directly with the naked eye.
Blood samples were collected from dogs that attended the National Taiwan University Veterinary Hospital, and signatures of agreement to participate in the study were obtained. Blood samples were submitted as part of routine tests. Complete blood counts were obtained using an Exigo veterinary hematology analyzer (Boule Diagnostics, Sweden). Genomic DNA was extracted from EDTA anti-coagulated blood using a Geneius Micro gDNA Extraction Kit (Geneaid Biotech, Taiwan). Multiplex PCR was performed using genomic DNA as the template. Microarray testing was then carried out to identify the MDR1 4-base deletion and SNP 180 genotypes in the tested dogs.
Dogs prescribed a P-glycoprotein-mediated medicine for chemotherapy, including chlorambucil, vinblastine, and vincristine [461415], were enrolled in this study. Cases in which the medicine dosage was not within the normal range [18] were excluded. Cases in which platelets showed clotting and the neutrophil number was above the normal range were also excluded. Side effects occurring after the first chemotherapy treatment, including neutropenia, thrombocytopenia, hypohemoglobin, diarrhea, and vomiting, were scored using the scoring system of the Veterinary Cooperative Oncology Group-Common Terminology Criteria for Adverse Events (VCOG-CTCAE) [921]. According to this scoring system, two treatment tolerance categories were defined: well-tolerated with a total score of 0 to 5 and adversely affected with a total score ≥ 6 [9]. High total scores indicated high adverse effects. The relationship between the genotypes of the two MDR1 gene loci to the side effect scores, and the genotypes relationship to the numbers of neutrophils and platelets and hemoglobin concentration were analyzed statistically by using one-way ANOVA. The genotypes relationship with dog breed was evaluated by using the Chi-squared test. GraphPad Prism (GraphPad Software, USA) was the statistical software used.
Separation of the 4-base no-deletion wild-type genotype (WW), 4-base deletion genotype (MM), and the hybrid genotype (WM) was tested by employing synthesized standard sequences with 5% agarose gel or 12% PAGE gel. The genotypes were barely detected using 5% agarose gel and could not be differentiated using 12% PAGE gel after 8 h of electrophoresis (panels A and B in Fig. 1).
The three SNP 180 genotypes (T/T, T/G, and G/G) could not be differentiated (panel C in Fig. 1) although simultaneous detection of 4-base and combined SNP 180 genotypes was possible when employing multiplex PCR with 2% agarose gel electrophoresis for 40 min (panel D in Fig. 1). Panel E in Fig. 1 shows the multiplex PCR results from three randomly-selected healthy dogs; the results indicate a failure to differentiate the three SNP 180 genotypes even with the multiplex PCR test.
The 4-base deletion and SNP 180 genotypes of MDR1 were clearly identified when using the oligonucleotide microarray with synthesized standard sequences (panel B in Fig. 2) or clinical samples of dog blood (panel C in Fig. 2). The results indicate that the oligonucleotide microarray was a much more practical and efficient approach, and could be an effective replacement for an electrophoresis and sequencing approach.
Genotypes of 100 randomly-chosen clinical blood samples from various dog breeds, including 5 collie and collie-related dogs, were tested using the oligonucleotide microarray. All showed wild-type homozygotes (WW) of the MDR1 4-base deletion. Among the MDR1 SNP 180 genotypes, 17 dogs were T/T, 63 were T/G, and 20 were G/G. The G allele variation frequency (%) was determined and no preference among dog breeds was detected in the SNP 180 genotypes (p = 0.4381; Table 3).
Twenty-eight dogs were included in a correlation analysis of the relationship between chemotherapy side effects and SNP 180 genotypes by applying the VCOG-CTCAE criteria [921]. The adverse side effect percentages for individuals with SNP 180 T/T, T/G, and G/G genotypes were 16.7%, 6.3%, and 0%, respectively (Table 4). This result demonstrates that the SNP 180 genotype can affect chemotherapy outcome. The SNP 180 G allele may induce more P-glycoprotein expression and pump more drugs out of the cells, resulting in fewer chemotherapy side effects. The SNP 180 genotype relationships with neutrophil number, thrombocyte number, and hemoglobin concentration were also analyzed. Thrombocyte number and hemoglobin concentration were not significantly different among the SNP 180 genotypes. However, the average neutrophil number increased progressively from the T/T genotype to the T/G and G/G genotypes (Fig. 3).
The traditional method for the detection of a 4-base deletion in the MDR1 gene employed high percentage agarose gel [10] or PAGE gel for prolonged electrophoresis [8]. Further advanced approaches have been developed, including amplification using allele-specific primers [219] and detection using allele-specific probes in real-time PCR [11]. However, they all still had detection problems. The SNP 180 polymorphisms of MDR1 gene could not be identified except through sequencing [1] or RFLP [17], and no integrated detection of both SNP 180 alleles and 4-base deletion mutations has been reported. Herein, we developed a simultaneous detection approach using an oligonucleotide microarray system. The accuracy of the approach was verified via sequencing both MDR1 gene loci of 28 dogs that were enrolled in a chemotherapy side effect study. The results showed that all of the microarray data correctly matched with the sequencing data producing an accuracy of 100% with no false positives or false negatives. The results for both genotypes were obtained simultaneously and within 4 h of test initiation. Thus, an oligonucleotide microarray-based system could provide a highly efficient method of identifying these two significant MDR1 genotypes, and the results could be available prior to initiating clinical treatment, preventing the occurrence of potential adverse effects related to treatment choice.
Among the 100 samples tested for 4-base deletion genotypes, all showed the homozygous wild-type, including the 5 collie dogs. This may be due to the low percentage of collies among the dogs sampled. Importation of a wild-type collie bloodline into Taiwan for breeding purposes might also have affected the results. Regardless, the three different SNP 180 genotypes were dispersed among the population. Unlike other studies [117], we found that the G allele variation in the SNP 180 genotype did not concentrate in collies. SNP 180 is located at intron 1 near the 5′ end of the gene where the more important promoter elements are located [20]. Previous authors have hypothesized that SNP 180 T>G variation at the promoter would cause P-glycoprotein over-expression, restricting the entry of drugs into the central nervous system [117], and facilitating drug elimination via urine and bile [7].
Dogs with the SNP 180 G/G genotype have shown a high potential for phenobarbital-resistance [117]. We therefore reasonably speculated that P-glycoprotein in the T/T genotype might give rise to high drug entry and low drug elimination, which could result in an increased risk of drug-related side effects. In this study, dogs with the SNP 180 T/T genotype had lower neutrophil numbers after chemotherapy than those in dogs with T/G and G/G genotypes, although there was no statistical significance to the differences (p = 0.3451). Dogs with SNP 180 T/T genotype displayed more diarrheas and vomiting after chemotherapy in this study, contributing to their high VCOG-CTCAE scores and adverse effect percentage, which surpassed the effects of neutrophil and thrombocyte numbers and hemoglobin concentration. Moreover, vincristine, one of the P-glycoprotein substrates, is metabolized via the MDR1 gene as well as via CYP3A4 and CYP3A5 in humans [12], making elucidation of the influence of side effects difficult.
Although we collected samples for more than one year, the number of dogs enrolled to evaluate the chemotherapy side effects was still small. In this study, we identified a distribution trend for two MDR1 genotypes (4-base deletion and SNP 180) in Taiwan and examined the correlation between three SNP 180 genotypes and the risk of a chemotherapy side effect. To increase the power of these initial results, a longer study with a larger sample size is necessary.
Figures and Tables
Fig. 1
Separation of MDR1 genotypes was tested by employing synthesized standard sequences and electrophoresis gels. (A and B) The 4-base deletion genotypes were detected by using 5% agarose gel and 12% polyacrylamide gel electrophoresis gel and 8 h electrophoresis, respectively. Lane M, marker; Lane 1, 4-base no-deletion wild-type genotype (WW genotype); Lane 2, 4-base deletion genotype (MM genotype); Lane 3, hybrid genotype (WM genotype); Lane 4, negative control. (C) The SNP 180 genotypes were detected by using 2% agarose gel and 40-min electrophoresis. Lane M, marker; Lane 1, T/T genotype; Lane 2, G/G genotype; Lane 3, T/G genotype; Lane 4, negative control. (D) Simultaneous detection of the 4-base deletion genotypes and SNP 180 genotypes of MDR1 using multiplex polymerase chain reaction (PCR). Lane M, marker; Lane 1, SNP 180 T/G genotype; Lane 2, 4-base deletion WM genotype; Lane 3, SNP 180 T/G and 4-base deletion WM genotypes; Lane 4, negative control. (E) Lane M, marker; Lanes 1–3, multiplex PCR results from three randomly-selected healthy dogs; Lane 4, negative control. Del 4, a 4-base deletion at exon 4 of the MDR1 gene at nucleotide position 230 (nt230[del4]); SNP, single nucleotide polymorphism.

Fig. 2
(A) Microarray map. VP1, positive control. The designation of each probe is shown in Table 2. Both the 4-base deletion and SNP 180 genotypes of MDR1 were clearly identified using an oligonucleotide microarray system and either the synthesized standard sequences (B) or clinical dog blood samples (C). WW, WM, and MM indicate wild-type homozygote, heterozygote, and homozygote 4-base deletion mutation genotype of MDR1, respectively. T/T, T/G, and G/G indicate allele-specific genotypes of SNP 180.

Fig. 3
The relevance of three SNP 180 genotypes to neutrophil number (A), thrombocyte number (B), and hemoglobin concentration (C) in blood from sampled dogs. SNP, single nucleotide polymorphism.
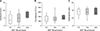
Table 1
Polymerase chain reaction primers designed for two loci amplification of the canine MDR1 gene

Table 2
Microarray probes designed for the simultaneous detection of the 4-base and SNP 180 genotypes of the canine MDR1 gene

Acknowledgments
This study was supported by the Ministry of Science and Technology of Taiwan (Grant No. 106-2622-B-002-010-CC2).
References
1. Alves L, Hülsmeyer V, Jaggy A, Fischer A, Leeb T, Drögemüller M. Polymorphisms in the ABCB1 gene in phenobarbital responsive and resistant idiopathic epileptic Border Collies. J Vet Intern Med. 2011; 25:484–489.


2. Baars C, Leeb T, von Klopmann T, Tipold A, Potschka H. Allele-specific polymerase chain reaction diagnostic test for the functional MDR1 polymorphism in dogs. Vet J. 2008; 177:394–397.


3. Borst P, Schinkel AH. P-glycoprotein ABCB1: a major player in drug handling by mammals. J Clin Invest. 2013; 123:4131–4133.


4. Bugde P, Biswas R, Merien F, Lu J, Liu DX, Chen M, Zhou S, Li Y. The therapeutic potential of targeting ABC transporters to combat multi-drug resistance. Expert Opin Ther Targets. 2017; 21:511–530.


5. Callaghan R, Luk F, Bebawy M. Inhibition of the multidrug resistance P-glycoprotein: time for a change of strategy? Drug Metab Dispos. 2014; 42:623–631.



6. Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, Hu T, Jiang L, Li J. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: a review of the past decade. Cancer Lett. 2016; 370:153–164.


7. Fromm MF. P-glycoprotein: a defense mechanism limiting oral bioavailability and CNS accumulation of drugs. Int J Clin Pharmacol Ther. 2000; 38:69–74.


8. Geyer J, Döring B, Godoy JR, Moritz A, Petzinger E. Development of a PCR-based diagnostic test detecting a nt230(del4) MDR1 mutation in dogs: verification in a moxidectin-sensitive Australian Shepherd. J Vet Pharmacol Ther. 2005; 28:95–99.


9. Gramer I, Kessler M, Geyer J. Determination of MDR1 gene expression for prediction of chemotherapy tolerance and treatment outcome in dogs with lymphoma. Vet Comp Oncol. 2015; 13:363–372.


10. Kawabata A, Momoi Y, Inoue-Murayama M, Iwasaki T. Canine mdr1 gene mutation in Japan. J Vet Med Sci. 2005; 67:1103–1107.

11. Klintzsch S, Meerkamp K, Döring B, Geyer J. Detection of the nt230[del4] MDR1 mutation in dogs by a fluorogenic 5′ nuclease TaqMan allelic discrimination method. Vet J. 2010; 185:272–277.


12. Lind DL, Fidel JL, Gay JM, Mealey KL. Evaluation of vincristine-associated myelosuppression in Border Collies. Am J Vet Res. 2013; 74:257–261.


13. Mealey KL, Bentjen SA, Gay JM, Cantor GH. Ivermectin sensitivity in collies is associated with a deletion mutation of the mdr1 gene. Pharmacogenetics. 2001; 11:727–733.


14. Mealey KL, Fidel J. P-glycoprotein mediated drug interactions in animals and humans with cancer. J Vet Intern Med. 2015; 29:1–6.



15. Mealey KL, Fidel J, Gay JM, Impellizeri JA, Clifford CA, Bergman PJ. ABCB1-1Δ polymorphism can predict hematologic toxicity in dogs treated with vincristine. J Vet Intern Med. 2008; 22:996–1000.


16. Mizukami K, Chang HS, Yabuki A, Kawamichi T, Hossain MA, Rahman MM, Uddin MM, Yamato O. Rapid genotyping assays for the 4-base pair deletion of canine MDR1/ABCB1 gene and low frequency of the mutant allele in Border Collie dogs. J Vet Diagn Invest. 2012; 24:127–134.


17. Mizukami K, Yabuki A, Chang HS, Uddin MM, Rahman MM, Kushida K, Kohyama M, Yamato O. High frequency of a single nucleotide substitution (c.-6-180T>G) of the canine MDR1/ABCB1 gene associated with phenobarbital-resistant idiopathic epilepsy in Border Collie dogs. Dis Markers. 2013; 35:669–672.



18. Plumb DC. Plumb's Veterinary Drug Handbook. Stockholm: PharmaVet;2011.
19. Stiedl CP, Weber K. Fast and simple detection methods for the 4-base pair deletion of canine MDR1/ABCB1 gene by PCR and isothermal amplification. J Vet Diagn Invest. 2017; 29:176–180.






 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download