Subjects
Twenty four men were enrolled at Asan Medical Center (Seoul, Korea). All enrolled subjects were healthy Korean male volunteers aged 19–50 years who weighed >50 kg and were within 20% of the ideal body weight. The platelet aggregation test was performed in healthy subjects who were not taking any medications and who did not have bleeding disorders. None of the subjects had significant cardiac, hepatic, renal, pulmonary, neurologic, gastrointestinal, or hematologic disorders as determined by medical history and physical examination. The physical examination included assessment of vital signs, electrocardiography, and clinical laboratory test (hematology, blood chemistry, and urinalysis). Subjects with a history of smoking or alcohol abuse, or those using any over-the-counter drug within 7 days before the first study day (Day 1) were excluded.
All laboratory tests other than the platelet aggregation test were performed at the Department of Laboratory Medicine, Asan Medical Center (accredited by the Korean Association of Quality Assurance for Clinical Laboratories). The platelet aggregation test was performed at the Clinical Pharmacology Lab of Asan Medical Center. The Institutional Review Board of Asan Medical Center approved the study protocol (registration No.; S2009-0248-0001), and all procedures were performed in accordance with the Good Clinical Practice guidelines[
6] and the Declaration of Helsinki and its amendments. All participants provided written, informed consent before the screening test for eligibility.
Pharmacodynamic plasma sample analyses (Platelet Aggregation Inhibition Assay)
The plasma assay for determining
in vitro platelet aggregation inhibition was performed using S- and R-indobufen (Ibustrin® tablet 200 mg) from Ildong Pharmaceuticals Co., Ltd. (Seoul, Korea). Each reference standard of S- and R-indobufen (0.5 g) was dissolved in methanol to generate a 10,000 mg/L solution, and then diluted using platelet-rich plasma (PRP) to produce 892 mixtures consisting of different concentrations of S- and R-indobufen. Basically, 0.25, 1, 2, 4, 8, 16, 24, 32, 64, and 128 mg/L of S-indobufen with corresponding 0 mg/L of R-indobufen, and vice versa were made using the plasma samples in each of 24 subjects. Then, the concentration combinations of S- and/or R-indobufen each that will be used in the experiment were randomly selected from 0.25, 1, 2, 4, 8, 16, 24, 32, 64, and 128 mg/L by generating random number using R software, and we conducted the experiment based on the preselected concentration combinations of S- and/or R-indobufen. The resultant final dataset used in this analysis consists of 366 and 364 “0” concentrations of S- and R-indobufen, and the other concentrations ranged from 36 and 48, respectively. Platelet aggregation inhibition was determined using a Four-Channel aggregometer (Chrono-Log 570VS Model, Chrono-Log Corp., Havertown, PA, USA) equipped with an AggroLink software package as described previously with modifications.[
7] In brief, PRP and platelet-poor plasma (PPP) were prepared by differential centrifugation (200 g for 10 min and 2,345 g for 10 min at 25℃, respectively). The PRP (0.3 mL) was incubated at 37℃ in the aggregometer for 5 min, followed by the addition of collagen (2 ug/mL) with continuous stirring. Platelet aggregation was recorded for up to 10 min and expressed as the maximal percentage change of light transmission from baseline using PPP as a reference, and maximal platelet aggregation (MPA) was calculated. Plasma samples from healthy male subjects were used for
in vitro assessments. No study drug was administered to the subjects.
Pharmacodynamic evaluation
A total of 892 MPA data with or without S- and R-Indobufen using plasma samples from 24 healthy male subjects were modeled using NONMEM® 7.2 with FOCE (first-order conditional estimation) and the INTERACTION method.[
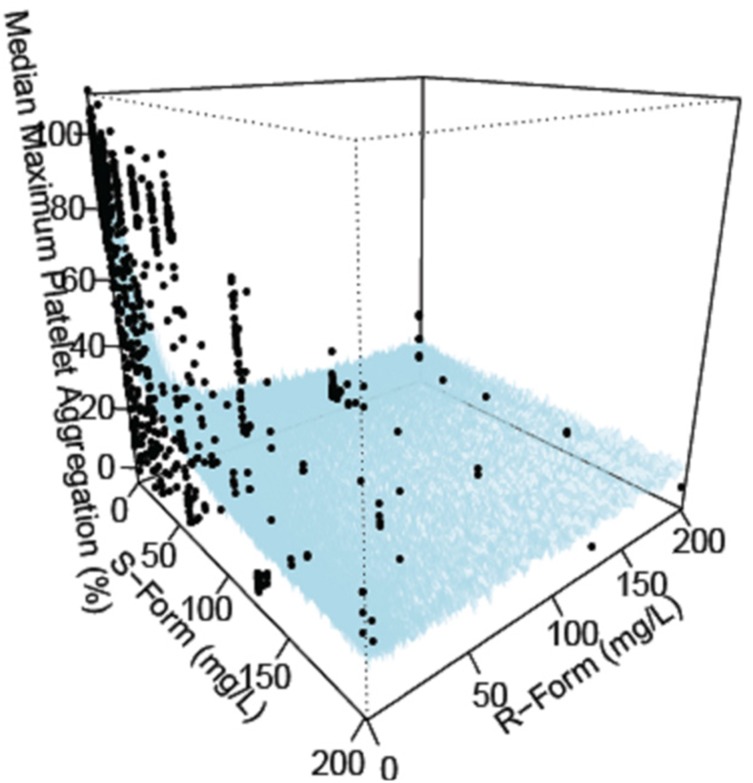
89] The baseline MPA measured without indobufen and MPA with indobufen were used as they are, considering the possible correlation between baseline MPA and the inhibitory effect of indobufen. The response surface model [
10] was used to characterize the pharmacodynamics of each enantiomer of indobufen including the interaction between enantiomers. First, the concentrations of R- (R
conc) and S-indobufen (S
conc) were normalized to the potency C50 as following UR and US:
Then, a family of drugs, each with a unique ratio of UR and US, was defined with an interaction term (ISR), and U50 was calculated as follows by applying fourth-order polynomial equation for empirical approximation and then simplifying the equation[
10]:
(equation 4 of the reference 10)
(equation 9 of the reference 10)
The concentration-response relation of the two stereoisomers was described using the inhibitory sigmoid I
max model [
11] as follows:
Base represents baseline MPA measured in plasma with neither S- nor R-indobufen; Imax represents the maximal inhibition, and gamma (γ) represents Hill's coefficient.
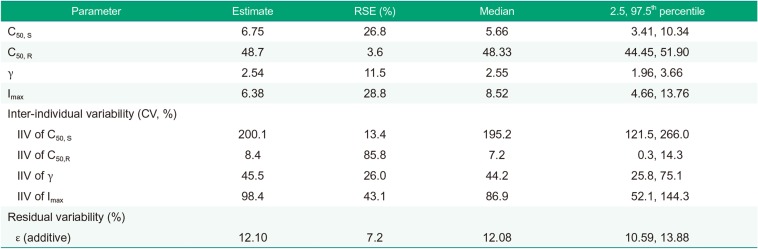
The inter-individual variability of parameters (C50, gamma, and Imax) was evaluated using an exponential model, whereas residual variability in platelet aggregation was most adequately described by an additive error model. The parameters for a specific subject were described using the following equation:
where PTV is the typical value of the parameter and ηi represents the normally distributed variables with a mean of zero (i.e., the unexplained inter-individual differences in a PD parameter among the individuals). Residual error was characterized using combined additive and proportional error models as described in the following equation:
where ε is zero-mean normally distributed variables.
Age, body weight, height, and platelet count were tested for their associations with various parameters. A likelihood ratio test was used to discriminate between hierarchical models at a p value of ≤0.05 because the distribution of −2 log likelihood of the models follows an approximate chi-square (χ2) distribution.
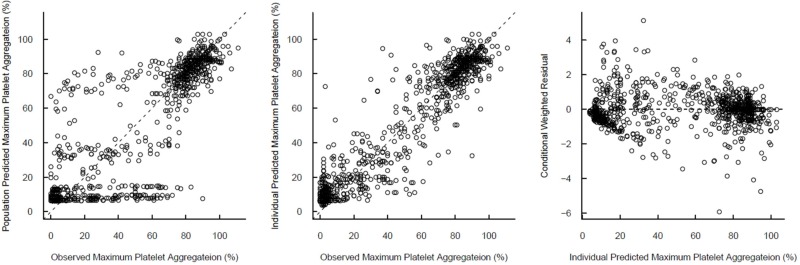
Standard diagnostic plots, including the observed values of the dependent variable (DV) versus the population predicted values (PRED) and the conditional weighted residuals (CWRES) versus PRED, were used for the diagnosis of optimum fit capabilities. Standard errors of parameter estimates of the PD model were used as a diagnostic. In the predictive check analysis, 1,000 datasets were simulated using NONMEM® 7.2 from the final PD model, and the median prediction was compared visually using the observed indobufen inhibition data in a response-surface plot. Bootstrapping, resampling technique with replacement, was performed to assess the bias and stability of the parameter estimates. A total of 1,000 bootstrap runs were performed, and the resulting parameter distributions were used to define 95% confidence intervals of the parameter estimates as 2.5th and 97.5th percentiles.
The modeling process was facilitated by the R statistical software (version 3.0.2, The R Foundation for Statistical Computing, Vienna, Austria, URL:
http://www.R-project.org).
Simulation study for the optimal dosage
After we characterized PD of each enantiomer we conducted simulation study predicting the treatment efficacy of each enantiomer to suggest optimal dosing regimens of each enantioselective formulation of indobufen which is potentially more adequate to use in clinical situations (
Fig. 1). Anti-platelet effect of each enantioselective indobufen was compared with the effect on 200 mg twice daily doses of
rac-indobufen, which is a current therapeutic doses of indobufen. Monte-carlo simulation was conducted to evaluate the concentration-anti-platelet effect relationships of each R- and S-indobufen formulation. Then, anti-platelet effect was simulated on 200 mg twice daily dosing regimen of each
rac-indobufen, S-indobufen, and R-indobufen using the average steady state concentration of S- and R-indobufen on 200 mg twice daily doses of
rac-indobufen in a previous clinical study.[
5]
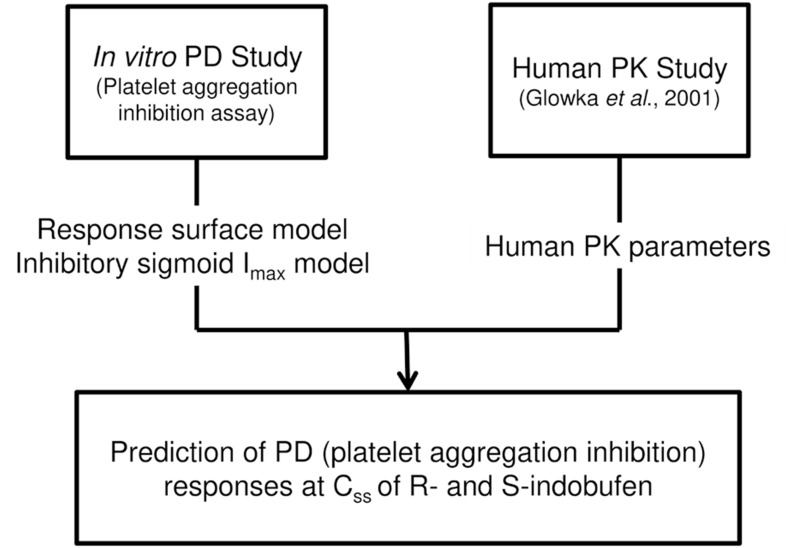 | Figure 1Schematic flow of the study design. Css, steady state concentration of R- and S-indobufen.
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print


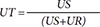
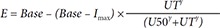

 XML Download
XML Download