MATERIALS AND METHODS
Data recruitment
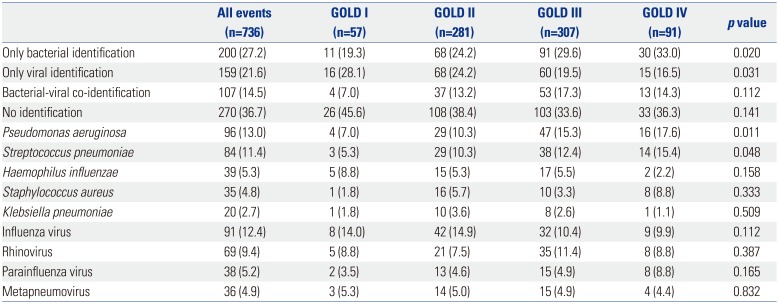
We reviewed and analyzed medical records of 736 cases of severe AECOPD at Korea University Guro Hospital from January 2011 to May 2017. We searched the electronic medical records database using the keywords “chronic obstructive pulmonary disease” and “acute exacerbation.” This study was approved by the Institutional Review Board of Korea University Guro Hospital (approved number: KUGH16131-002).
COPD was diagnosed based on the guidelines of the Global Initiative for Chronic Obstructive Lung Disease (GOLD). AECOPD was defined as “an acute event characterized by a worsening of the patient's respiratory symptoms that is beyond normal day-to-day variation and leads to a change in medication.”
1314 All patients had a previous pulmonary function test, and a ratio of forced expiratory volume in one second (FEV
1) to forced vital capacity (FVC) of <70%. All patients were >40 years old.
Patients were excluded if 1) the cause of admission was something other than AECOPD, such as heart failure, arrhythmia, acute pulmonary edema, acute pulmonary embolism, or pneumothorax; 2) they had a comorbidity that can affect immunity, such as cancer or autoimmune disease; 3) no laboratory or culture test was conducted within 24 hr of hospitalization; or 4) they exhibited mild to moderate exacerbation of COPD.
Laboratory tests
All cultures, antigen detection assays, and polymerase chain reaction (PCR) assays of blood, sputum, urine, or naso- or oro-pharyngeal swab samples were obtained by a physician within 24 hr of admission.
Bacterial cultures were performed on blood and sputum samples using standard techniques. Urinary antigen testing was performed to detect Streptococcus pneumoniae. Real-time PCR (RT-PCR) assays were performed using naso- or oro-pharyngeal swab samples to detect the following respiratory viruses: influenza virus, respiratory syncytial virus, parainfluenza virus, coronavirus, rhinovirus, enterovirus, adenovirus, bocavirus, and metapneumovirus. A rapid antigen test was used to test to detect influenza.
Pathogen identification
A bacterial pathogen was determined to be present if 1) gram-positive or gram-negative bacteria were detected in a blood or sputum sample or if 2) Streptococcus pneumoniae was detected in urine by an antigen detection assay.
A viral pathogen was determined to be present if 1) one of the viruses listed above was detected in a naso- or oro-pharyngeal swab by an RT-PCR assay or if 2) influenza virus was detected by a rapid antigen test.
Statistical analysis
Data were analyzed using SPSS 20 software (IBM Corp., Armonk, NY, USA). Data were presented as mean±standard deviation or number and percent of each group. When events were classified by GOLD stage or month, we used a one-way analysis of variance. When events were classified by in-hospital mortality, chi-square test or Fisher's test was used for discrete data, and an independent t-test or Mann-Whitney test was used for continuous data. All p-values <0.050 were considered statistically significant.
DISCUSSION
This was a large, retrospective, population-based, single center study of bacterial and viral pathogens associated with AECOPD in the Republic of Korea, including epidemiological factors. Bacterial and viral infections are the most common causes of AECOPD.
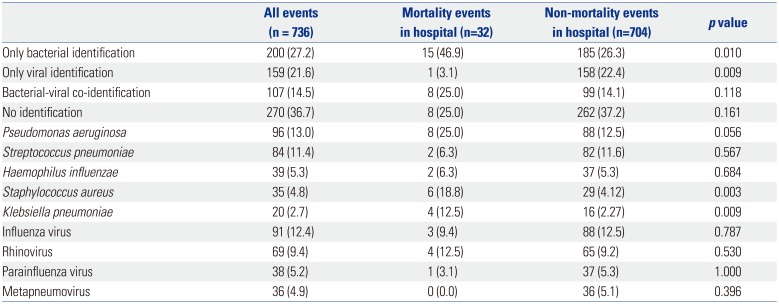
7 The bacterial identification rate in AECOPD events has previously been reported to be 40–60% and the viral identification rate 20–40%.
9121516 Our study showed similar results, with a 41.7% bacterial identification rate and 36.1% viral identification rate.
The most commonly identified bacteria in previous studies of AECOPD have been
H. influenzae,
Moraxella catarrhalis, and
S. pneumoniae.
15 However, in our study,
M. catarrhalis was identified in only 8 (1.1%) of 736 AECOPD events. Along with
H. influenzae and
S. pneumoniae, in our study,
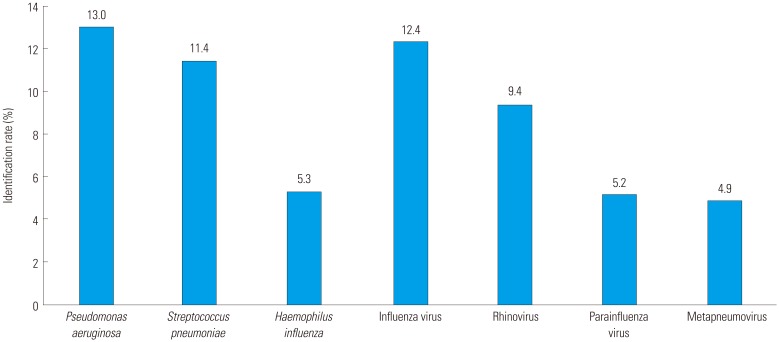
P. aeruginosa was identified as a major AECOPD pathogen. Previous studies have found that
P. aeruginosa identification rates increase with GOLD stage and that higher
S. pneumoniae identification rates are associated with comorbidity.
17 Our results agree with these previous studies: we found that
P. aeruginosa identification increased as the GOLD stage increased. The correlation between
S. pneumoniae identification rate and GOLD stage seems to be due to high comorbidity at more advanced GOLD stages. The low identification rate of
M. catarrhalis may be due to regional differences. For other reasons,
M. catarrhalis may not be cultured better than other bacteria, so the identification rate may be low.
New molecular diagnostic methods, such as nucleic acid amplification tests, antigen detection assays, and PCR assays, have revealed the importance of viral infection as a cause of AECOPD. Meanwhile, virus has also been identified as a major cause of exacerbation in AECOPD patients with low C-reactive protein levels.
18 Respiratory viral infections affect epithelial cells in the airway and lead to microvascular dilatation, edema, inflammatory cell infiltration, and reduction of mucociliary clearance.
19 In previous studies of AECOPD, rhinovirus was the most commonly identified virus.
920 In our study, influenza and parainfluenza viruses along with rhinovirus had the highest identification rates. The high influenza identification rates in our study are likely due to influenza epidemics in Korea in 2014 and 2015; 51 of the 91 events with influenza virus identification occurred during this time. These results suggest that rhinovirus is the virus most commonly identified with AECOPD unless there is another virus outbreak.
As mentioned above,
P. aeruginosa is frequently identified in severe and very severe COPD patients. This is thought to be related to bronchiectasis. A high prevalence of bronchiectasis was found in patients with moderate to severe COPD.
21 Chest computed tomography was performed in 526 of the 736 events, and bronchiectasis was diagnosed in 222 events. The prevalences of bronchiectasis were 10.3% for GOLD stage I, 36.9% for stage II, 49.0% for stage III, and 54.1% for stage IV. When choosing antibiotics for use in patients with severe or very severe AECOPD, antibiotics with antimicrobial activity against
P. aeruginosa should be considered.
The prevalence of respiratory viruses is closely related to temperature and humidity.
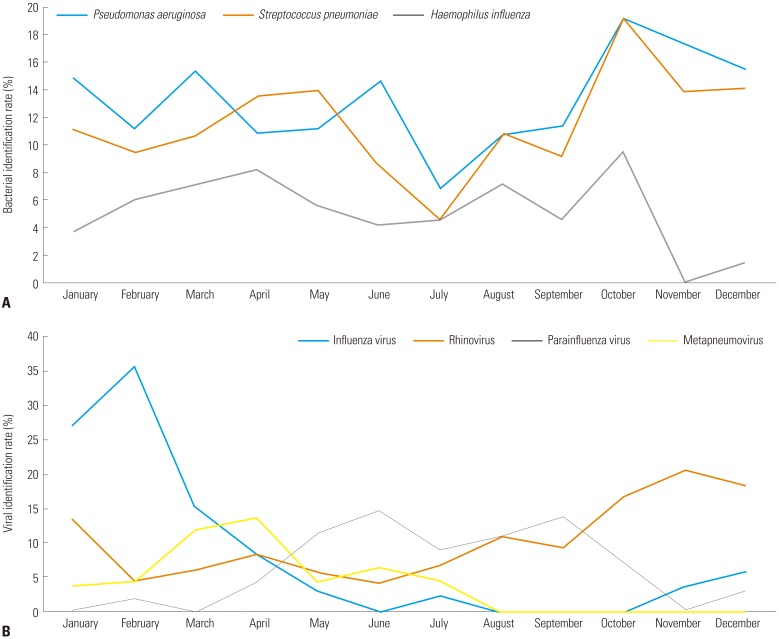
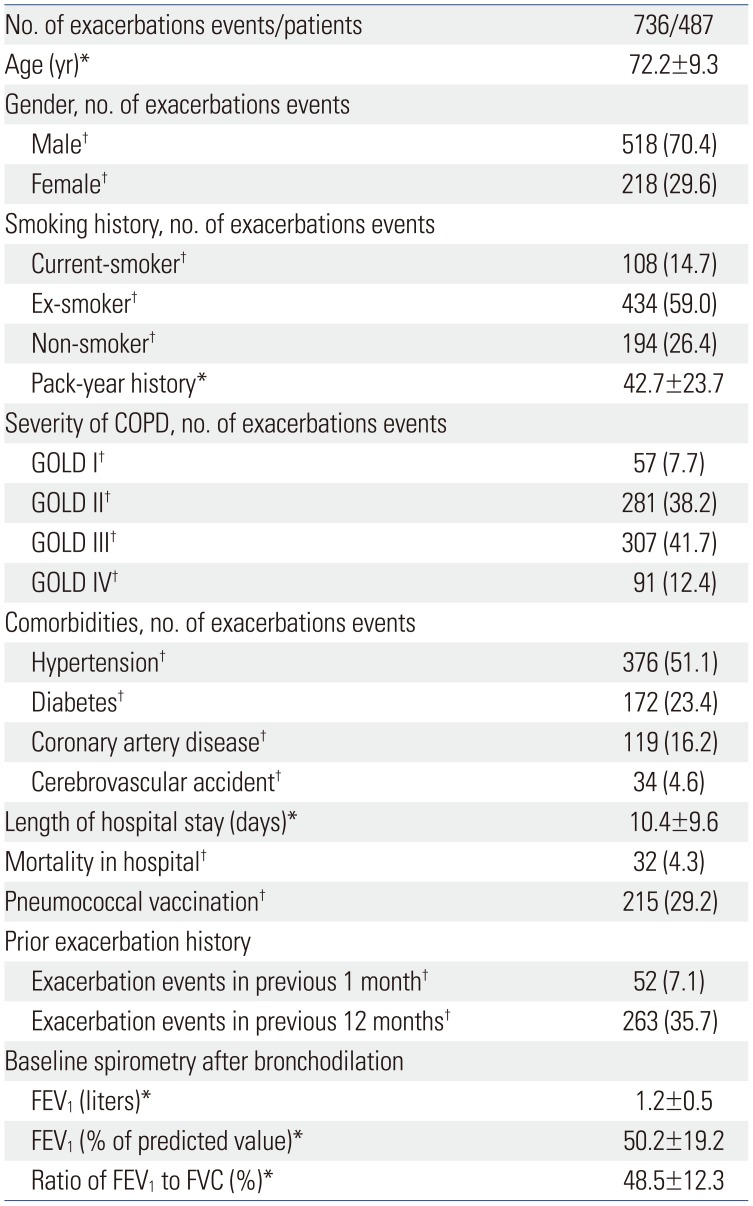
2223 Korea has four distinct seasons, and worldwide, viral identification in AECOPD has shown seasonal variation. In our study, influenza virus and rhinovirus were detected in the winter, metapneumovirus in the spring and parainfluenza virus in the summer. Our results are consistent with a previous study conducted in Istanbul, in which metapneumovirus was prominent in the spring and parainfluenza virus was predominant in the summer.
24 Knowledge of seasonal virus prevalence will help in prevention and treatment of respiratory viral infections in AECOPD.
The rate of mortality due to AECOPD was higher when bacteria were identified. Age, comorbidity, and home oxygen use were higher in the mortality group than in the non-mortality group. Therefore, these factors (the type of infection and the basal condition of the patient) should be taken into consideration when predicting prognosis and mortality.
When there is co-identification of bacterial and viral infection, it is not known which infection occurs first. Recent studies have shown that respiratory viral infections cause dysregulation of iron homeostasis mechanisms associated with secondary bacterial infections.
25 In addition, respiratory syncytial virus promotes development of
P. aeruginosa biofilms
in vitro and
in vivo. On the other hand, bacterial infections in the airway can promote respiratory viral infection.
26 When the lungs are infected with
P. aeruginosa, the tight junction between epithelium is damaged resulting in release of lactate dehydrogenase, destroying the lung barrier.
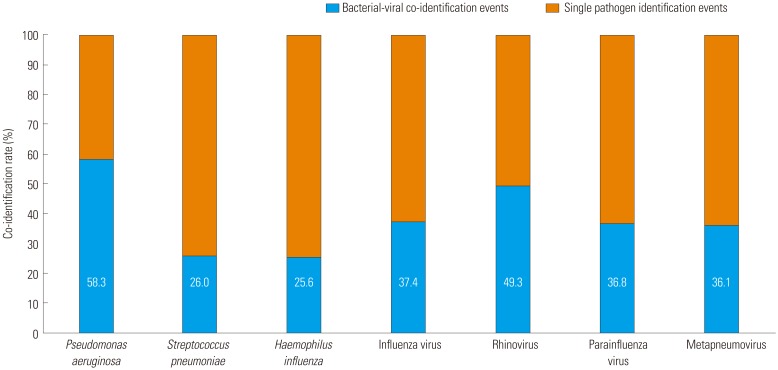
2728 Of the bacteria in our study,
P. aeruginosa was most commonly co-identified with a virus. Further studies on the interactions between
P. aeruginosa and viruses are needed.
Our study has some limitations. First, it is a retrospective, single center study. Therefore, many events were excluded through lack of data and exclusion criteria. For example, we evaluated pneumococcal vaccination, but did not find influenza vaccination coverage for all patients. Thus, we could not analyze the effect of influenza vaccination at the time of the influenza epidemic. Also, chest computed tomography was performed only in some cases, so it was not applicable to all events. Second, colonization could not be distinguished in the analysis of detection; in particular, it would have been advantageous if there had been previous or follow-up data from sputum culture. Third, influenza rapid antigen test was performed only in 247 (33.6%) events. Additionally, it was mainly performed only in the epidemic season. We think this would have affected the analysis of seasonal variation. Fourth, the phenotype of COPD might have a major impact on pathogen identification. However, through retrospective data, it was difficult to accurately identify COPD phenotype. Thus, we could not analyze according to COPD phenotype. However, this study has merits: it is a large study that evaluates bacterial and viral identification rates associated with AECOPD in South Korea, and it provides information to help understand the epidemiology of AECOPD.
In conclusion, bacterial and viral infections are a major cause of AECOPD. The prevalence of identified pathogens has regional variability. In our study, the most commonly identified bacterium associated with AECOPD was P. aeruginosa, and the most commonly identified virus was influenza. M. catarrhalis showed a low identification rate, which is characteristic of Korea. Viruses were shown to be prevalent in certain seasons in Korea. Understanding the epidemiology of infections in AECOPD will help in selecting antibiotics and treatment.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download