The diagnosis of acute myeloid leukemia (AML) has evolved over the past 15 years into a disease characterization technique that is largely based on cytogenetic and molecular analysis. over the past 15 years into a disease characterization technique that is largely based on cytogenetic and molecular analysis. Since the WHO classification was first proposed, it has been established as a formal tool for the diagnosis of hematologic malignancies [
1]. The 2016 WHO classification incorporated new information that has emerged since the previous 2008 WHO classification, including acceptance of previous provisional entities such as AML with mutated
NPM1 and AML with double
CEBPA mutations as definite ones [
2]. However, this classification system must be contemporary and match with the rate of accumulating evidence; thus, there has been pressure to revise the AML classification after a certain period by collecting data and assessing their relevance. From a diagnostic perspective, application of the new criteria should be preceded by an understanding of the expected changes, accompanied by a revised classification. We reclassified our AML database that was created according to the 2008 WHO classification, and estimated the changes in data distribution and position on application of the 2016 WHO classification. Particularly, we focused on the newly introduced mutations, including
NPM1 and
CEBPA.
We reviewed 610 patients who were diagnosed as having AML and were treated at Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea between January 2014 and June 2017. This study was approved by the Institutional Review Board (IRB) of St. Mary's Hospital affiliated with The Catholic University of Korea (IRB No: KC18RESI0227). Patient characteristics according to the 2016 WHO classification are summarized in
Supplemental Data Table S1. The patients included 335 males and 275 females with a median age of 50 years (range, 1–88 years). Patient bone marrow aspirates and biopsy samples were reviewed independently by three hematopathologists. The patients' medical records, including history of chemotherapy, were reviewed. Cytogenetic abnormalities were classified according to the 2016 International System for Human Cytogenetic Nomenclature guidelines [
3]. Multiplex reverse transcriptase-PCR was performed to detect the presence of
RUNX1/RUNX1T1,
CBFB/MYH11,
PML/RARA,
MLLT3/KMT2A,
DEK/NUP213, and
BCR/ABL1 using a HemaVision kit (Bio-Rad Laboratories, Hercules, CA, USA). Mutations in the
CEBPA and
NPM1 genes were analyzed by bidirectional Sanger sequencing using primers designed through Primer3 (
http://bioinfo.ut.ee/primer3/) and described based on reference to GenBank sequences (
CEBPA, NM_004364.4;
NPM1, NM_002520.6).
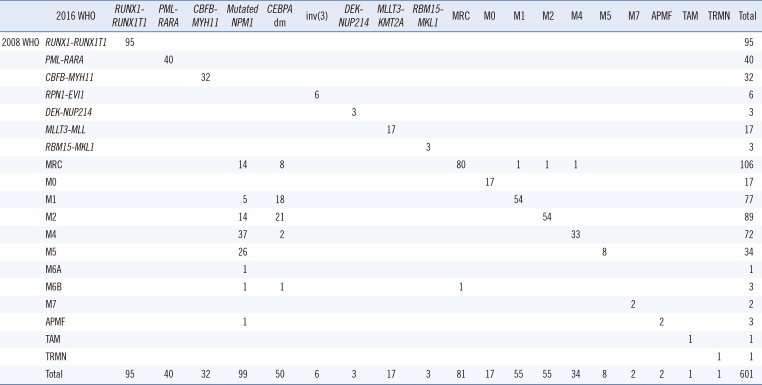
Seven (1.1%) patients were categorized as having myelodysplastic syndrome (MDS) with excess blasts, because the definition of myeloid neoplasms with erythroid predominance was modified by shifting the main criteria for calculating blast percentage from non-erythroid cells to all nucleated marrow cells. Two patients were classified as having myeloid neoplasms with germline predisposition because they harbored a germline
CEBPA mutation. AML with recurrent genetic abnormalities accounted for 57.4% (345/601) based on the 2016 WHO classification. AML with mutated
NPM1 was the most common form, followed by AML with
RUNX1-RUNX1T1, AML with double
CEBPA mutations, and AML with
CBFB-MYH11 (5.3%, N=32) (
Fig. 1). Among AML with myelodysplasia-related changes (MRC) patients, seven (8.6%) had a history of MDS or myelodysplastic/myeloproliferative neoplasm, and 40 (49.4%) had MDS-related cytogenetic abnormalities. Because del(9q) was removed as a defining cytogenetic abnormality for AML-MRC, 10 patients with del(9q) were removed from the AML-MRC group; six of these patients were moved to AML with double
CEBPA mutations and one diagnosis was changed to AML with mutated
NPM1, while the others were reclassified as AML, not otherwise specified (NOS) (
Table 1).
Fig. 1
Distribution of subtypes in AML patients classified according to 2016 WHO classification.
Abbreviations: AML, acute myeloid leukemia; N, number of patients (%); dm, double mutation; MRC, AML with myelodysplasia-related changes; M0, AML with minimal differentiation; M1, AML without maturation; M2, AML with maturation; M4, acute myelomonocytic leukemia; M5, acute monoblastic and monocytic leukemia; APMF, acute panmyelosis with myelofibrosis; TAM, transient abnormal myelopoiesis associated with Down syndrome; TRMN, therapy-related myeloid neoplasms.
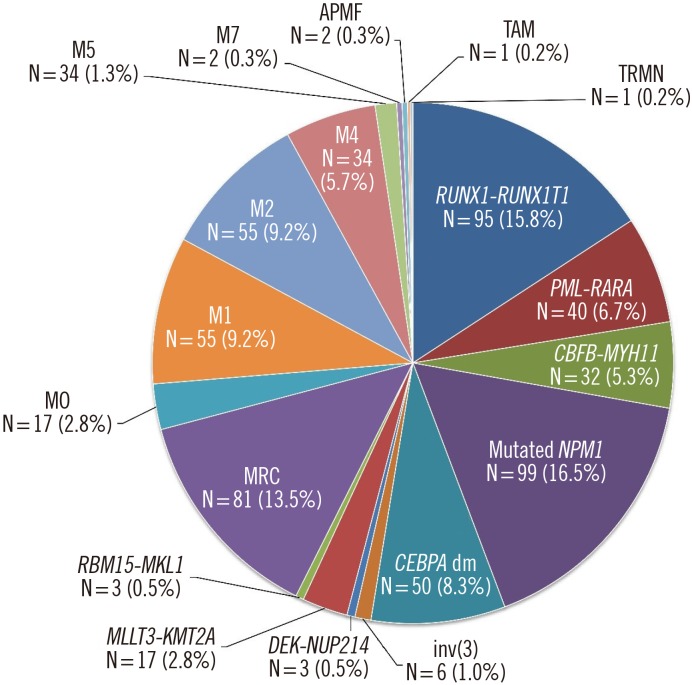

Table 1
Comparison of the distribution of AML patients based on the 2008 and 2016 WHO classifications
|
2016 WHO |
RUNX1-RUNX1T1
|
PML-RARA
|
CBFB-MYH11
|
Mutated NPM1
|
CEBPA dm |
inv(3) |
DEK-NUP214
|
MLLT3-KMT2A
|
RBM15-MKL1
|
MRC |
M0 |
M1 |
M2 |
M4 |
M5 |
M7 |
APMF |
TAM |
TRMN |
Total |
|
2008 WHO |
RUNX1-RUNX1T
|
95 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
95 |
|
PML-RARA
|
|
40 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
40 |
|
CBFB-MYH11
|
|
|
32 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
32 |
|
RPN1-EVI1
|
|
|
|
|
|
6 |
|
|
|
|
|
|
|
|
|
|
|
|
|
6 |
|
DEK-NUP214
|
|
|
|
|
|
|
3 |
|
|
|
|
|
|
|
|
|
|
|
|
3 |
|
MLLT3-MLL
|
|
|
|
|
|
|
|
17 |
|
|
|
|
|
|
|
|
|
|
|
17 |
|
RBM15-MKL1
|
|
|
|
|
|
|
|
|
3 |
|
|
|
|
|
|
|
|
|
|
3 |
|
MRC |
|
|
|
14 |
8 |
|
|
|
|
80 |
|
1 |
1 |
1 |
|
|
|
|
|
106 |
|
M0 |
|
|
|
|
|
|
|
|
|
|
17 |
|
|
|
|
|
|
|
|
17 |
|
M1 |
|
|
|
5 |
18 |
|
|
|
|
|
|
54 |
|
|
|
|
|
|
|
77 |
|
M2 |
|
|
|
14 |
21 |
|
|
|
|
|
|
|
54 |
|
|
|
|
|
|
89 |
|
M4 |
|
|
|
37 |
2 |
|
|
|
|
|
|
|
|
33 |
|
|
|
|
|
72 |
|
M5 |
|
|
|
26 |
|
|
|
|
|
|
|
|
|
|
8 |
|
|
|
|
34 |
|
M6A |
|
|
|
1 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1 |
|
M6B |
|
|
|
1 |
1 |
|
|
|
|
1 |
|
|
|
|
|
|
|
|
|
3 |
|
M7 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
2 |
|
|
|
|
2 |
|
APMF |
|
|
|
1 |
|
|
|
|
|
|
|
|
|
|
|
2 |
|
|
|
3 |
|
TAM |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1 |
|
|
1 |
|
TRMN |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
1 |
|
1 |
|
Total |
95 |
40 |
32 |
99 |
50 |
6 |
3 |
17 |
3 |
81 |
17 |
55 |
55 |
34 |
8 |
2 |
2 |
1 |
1 |
601 |

We investigated differences in hematologic variables according to the 2016 WHO classification using a one-way analysis of variance followed by the Bonferroni post hoc test. We used SPSS 12.0.1 for Windows (SPSS Inc., Chicago, IL, USA) and considerd
P≤0.05 (two-sided) statistically significant. Patient age was the highest in the groups AML with mutated
NPM1 and AML-MRC, but lowest in the group AML with
RUNX1-RUNX1T1 (
P<0.001). Leukocyte count was higher in AML with mutated
NPM1 than in AML with
RUNX1-RUNX1T1 and AML with
PML-RARA (
P=0.005). The blast percentage in peripheral blood was the highest in AML with double
CEBPA mutations, which was greater than that in AML with
RUNX1-RUNX1T1 or
PML-RARA (
P<0.001). However, there was no significant difference in the Hb or platelet count (
Supplemental Data Fig. S1).
The 2016 WHO classification newly defined
NPM1 and double
CEBPA mutations for diagnostic classification. Therefore, we evaluated the previous diagnoses of these two new classifications. AML with mutated
NPM1 was the most common form of AML in our database (N=99, 16.5%). It was notable that all the patients in this group were over 18 years old. The majority of the patients in the AML with mutated
NPM1 group had previously been diagnosed as having AML-NOS, including acute myelomonocytic leukemia (N=37), acute monoblastic/monocytic leukemia (N=26), AML with maturation (N=14), AML without maturation (N=5), acute erythroid leukemia (N=2), and acute panmyelosis with myelofibrosis (N=1), according to the 2008 WHO classification. Others (N=14) had been diagnosed as having AML-MRC showing only dyspoiesis without MDS-related cytogenetic abnormalities. The association of monocytic differentiation and
NPM1 mutation was consistent with previous results [
45].
NPM1 mutation in exon 12 was the most frequently detected mutation in cytogenetically normal AML patients. This is supported by our observation that 93 (93.9%) patients diagnosed as having AML with mutated
NPM1 showed a normal karyotype. Five of the six patients with an abnormal karyotype showed mosaicism with a normal karyotype, which can be explained as an acquired secondary event [
6]. The characteristics of
NPM1 mutation (N=102) are summarized in
Supplemental Data Fig. S2: type A mutation was predominant, followed by type B, type D, and type I, whereas type C, type J, type N, type R, and four individual mutations (c.863_864insTAAA, c.863_864insTTTG, c.864_876delinsCCAAGATCTCTGGCATT, c.869_873delinsCCTTGGCTC) were detected in one patient each.
There were 77 (12.8%) patients with
CEBPA mutations in our database, including 46 (59.7%) with double
CEBPA mutations. Four of these patients (5.2%) had one homozygous mutation, two patients had three mutations, and the other 25 (32.5%) had a monoallelic mutation. Thus, AML with double
CEBPA mutations accounted for 8.3% of all forms of AML in the database. Previous classifications of AML-NOS (N=50), including AML with maturation (N=21), AML without maturation (N=18), acute myelomonocytic leukemia (N=2), acute erythroid leukemia (N=1), and AML-MRC (N=8) were reclassified as AML with double
CEBPA mutations according to the 2016 WHO classification. Thirty-eight (76.0%) of these 50 patients had a normal karyotype, and six patients with del(9q) were also classified in this group, which is recognized as a cytogenetic abnormality for AML-MRC according to the 2008 WHO classification. Previous studies demonstrated that the majority of patients with del(9q) were diagnosed as having AML with or without maturation and a normal karyotype [
78]. In addition, these patients tended to have an earlier age of onset, higher Hb levels, and lower platelet counts than patients with other AML classifications, although the differences were not statistically significant [
9]. Moreover, patients with double
CEBPA mutations were reported to show a homogeneous gene expression profile and a favorable clinical outcome [
1011].
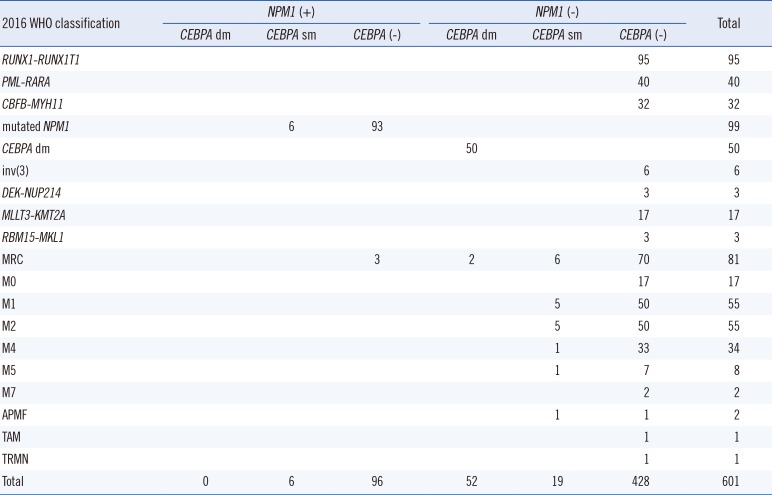
Interestingly, the two new classifications of
NPM1 and double
CEBPA mutations were mutually exclusive without overlapping with any other form of AML with recurrent genetic abnormalities (
Table 2). There were five patients with
NPM1 mutations (N=3) or double
CEBPA mutations (N=2) who were classified as having AML-MRC because of the diagnostic precedence of MDS-related cytogenetic abnormalities. AML with mutated
NPM1 became the most common AML in the 2016 WHO classification. Although approximately 80% of
NPM1 mutations identified were type A, other types cannot be ignored.
Table 2
Distribution of NPM1 and CEBPA mutations in 2016 WHO classified AML patients
|
2016 WHO classification |
NPM1 (+)
|
NPM1 (−) |
Total |
|
CEBPA dm |
CEBPA sm |
CEBPA (−) |
CEBPA dm |
CEBPA sm |
CEBPA (−) |
|
RUNX1-RUNX1T1
|
|
|
|
|
|
95 |
95 |
|
PML-RARA
|
|
|
|
|
|
40 |
40 |
|
CBFB-MYH11
|
|
|
|
|
|
32 |
32 |
|
mutated NPM1
|
|
6 |
93 |
|
|
|
99 |
|
CEBPA dm |
|
|
|
50 |
|
|
50 |
|
inv(3) |
|
|
|
|
|
6 |
6 |
|
DEK-NUP214
|
|
|
|
|
|
3 |
3 |
|
MLLT3-KMT2A
|
|
|
|
|
|
17 |
17 |
|
RBM15-MKL1
|
|
|
|
|
|
3 |
3 |
|
MRC |
|
|
3 |
2 |
6 |
70 |
81 |
|
M0 |
|
|
|
|
|
17 |
17 |
|
M1 |
|
|
|
|
5 |
50 |
55 |
|
M2 |
|
|
|
|
5 |
50 |
55 |
|
M4 |
|
|
|
|
1 |
33 |
34 |
|
M5 |
|
|
|
|
1 |
7 |
8 |
|
M7 |
|
|
|
|
|
2 |
2 |
|
APMF |
|
|
|
|
1 |
1 |
2 |
|
TAM |
|
|
|
|
|
1 |
1 |
|
TRMN |
|
|
|
|
|
1 |
1 |
|
Total |
0 |
6 |
96 |
52 |
19 |
428 |
601 |

Next-generation sequencing (NGS) has recently replaced Sanger sequencing as the main method to identify common and significant mutations simultaneously, with excellent detection power [
12]. The other newly included classification, AML with double
CEBPA mutations, was the third most frequent form of AML with recurrent genetic abnormalities. Detection of
CEBPA mutation is hampered mainly by the high GC content of this gene. Although more efficient and sensitive techniques such as NGS are emerging to address this challenge, Sanger sequencing still plays an important role in clinical
CEBPA testing [
13].
In summary, we found that implementation of the updated 2016 WHO classification of AML would not pose major difficulties in clinical practice and could help reduce the rate of ambiguous diagnoses for a more accurate prognosis. Hematopathologists should review and prepare genetic tests that are essential for the new classification, according to their clinical laboratory conditions. Future studies incorporating prognosis in the 2016 WHO classification would help provide baseline data for upgrading the classification to achieve better risk stratification and patient-oriented therapeutic strategies.







 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download