Clostridium difficile infection (CDI) has become the most common cause of healthcare-associated diarrhea, with an increasing prevalence in high-income countries [
1234]. In the United States,
C. difficile is the most frequently reported nosocomial pathogen. The incidence of CDI has increased from 4.5 per 1,000 adult discharges in 2001 to 8.2 per 1,000 adult discharges in 2010. Patients with CDI have higher health care costs: annual attributable costs exceed $1.5 billion in the United States [
2]. In Korea, a nationwide study revealed that total incidence of CDI has increased significantly from 1.7 per 1,000 adult admissions in 2004 to 2.7 per 1,000 adult admissions in 2008 [
5].
Rapid and accurate diagnosis of CDI is crucial for patient care, infection control, and surveillance. Various assays are currently available for diagnosing CDI, including the cell cytotoxicity neutralization assay (CCNA), toxigenic culture (TC), toxin AB enzyme immunoassay (toxin AB EIA), glutamate dehydrogenase (GDH) assay, and nucleic acid amplification tests (NAATs). Algorithmic approaches have also been developed to improve the diagnostic performance, and several guidelines for CDI diagnosis have been established [
5678]. However, this wide variation in approaches has hindered universal application of these guidelines. Moreover, there is currently no consensus for the best CDI diagnostic assay or strategy to adopt in Korea. As a first step toward standardization of CDI diagnosis in Korea, we conducted a national survey to investigate the diagnostic assays for CDI used in clinical laboratories.
In May 2015, we administered a voluntary online survey on laboratory diagnosis for CDI to health professionals in 120 clinical microbiology laboratories (
https://docs.google.com/forms/u/0/). Questions covered the current assays used for CDI diagnosis, including the toxin AB EIA, NAAT,
C. difficile culture, GDH assay, and CCNA, and the number of examined specimens. This study was approved by the Institutional Review Board of Inje University Sanggye Paik Hospital (IRB No. SGPAIK-2018-10-010), which waived the requirement for informed consent. The data was organized and analyzed using Microsoft Excel 2016 (Microsoft, Redmond, WA, USA). Statistical analysis was performed using MedCalc Version 10.0 (MedCalc Software bvba, Ostend, Belgium). The Mann-Whitney test was used to compare the number of examined specimens between assays.
P<0.05 was considered statistically significant.
Responses were obtained from 66 laboratories, including 61 hospitals (number of beds≥1,000, N=11; 500–1,000, N=42; 300–500, N=3; <300, N=5) and five commercial laboratories (CL). The 61 hospitals were located in 6 metropolitan cities (Seoul, N=26; Incheon, N=4; Daegu, N=4; Busan, N=3, Gwangju, N=3; Daejeon, N=1; Ulsan, N=1) and 5 provinces (Gyeonggi, N=12; Chungbuk, N=2; Gyeongnam, N=2; Jeonbuk, N=2; Jeonnam, N=1). Among them, nine laboratories reported having not conducted any CDI assay. All hospitals with ≥1,000 beds performed CDI assays, whereas 88.1% (37/42) of hospitals with 500–1,000 beds and 50.0% (4/8) of hospitals with <500 beds (including the 300–500 and <300 beds categories) performed CDI assays.
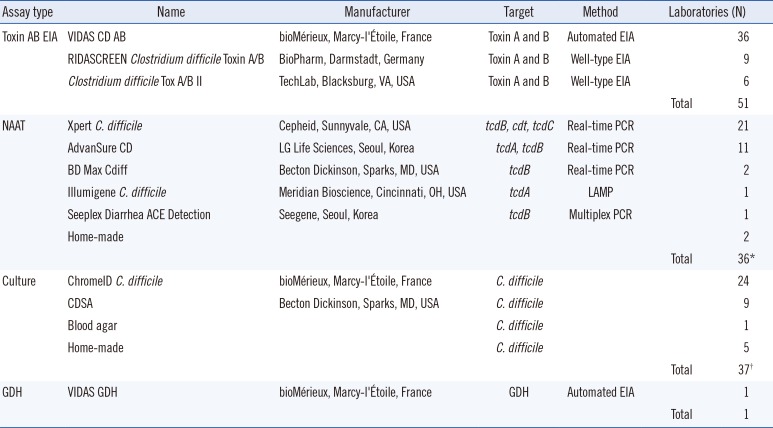
The various assay methods used in the participating laboratories are summarized in
Table 1. The toxin AB EIA was the most popular assay. Among the 57 laboratories that reported performing CDI assays, 51 (89.5%) used the toxin AB EIA, either alone or in combination with other assays. NAATs and
C. difficile culture, alone or in combination with other assays, were used in 37 (64.9%) laboratories. Only one laboratory used the GDH assay, which was conducted in combination with the toxin AB EIA. Forty-five (78.9%) laboratories used more than one assay. However, no laboratory reported performing the CCNA.
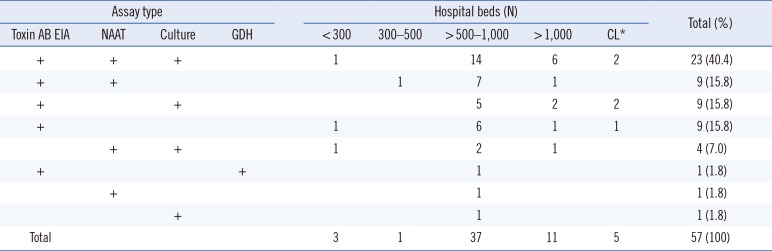
Table 2 shows combinations of assay types for diagnosis of CDI according to the size of hospitals. Assay type (single or combination) did not significantly differ by hospital size.
Table 1
Clostridium difficile assay methods and the numbers of laboratories that participated in the survey
|
Assay type |
Name |
Manufacturer |
Target |
Method |
Laboratories (N) |
|
Toxin AB EIA |
VIDAS CD AB |
bioMérieux, Marcy-l'Étoile, France |
Toxin A and B |
Automated EIA |
|
36 |
|
RIDASCREEN Clostridium difficile Toxin A/B |
BioPharm, Darmstadt, Germany |
Toxin A and B |
Well-type EIA |
|
9 |
|
Clostridium difficile Tox A/B II |
TechLab, Blacksburg, VA, USA |
Toxin A and B |
Well-type EIA |
|
6 |
|
|
|
|
Total |
51 |
|
NAAT |
Xpert C. difficile
|
Cepheid, Sunnyvale, CA, USA |
tcdB, cdt, tcdC
|
Real-time PCR |
|
21 |
|
AdvanSure CD |
LG Life Sciences, Seoul, Korea |
tcdA, tcdB
|
Real-time PCR |
Total |
11 |
|
BD Max Cdiff |
Becton Dickinson, Sparks, MD, USA |
tcdB
|
Real-time PCR |
|
2 |
|
Illumigene C. difficile
|
Meridian Bioscience, Cincinnati, OH, USA |
tcdA
|
LAMP |
|
1 |
|
Seeplex Diarrhea ACE Detection |
Seegene, Seoul, Korea |
tcdB
|
Multiplex PCR |
|
1 |
|
Home-made |
|
|
|
|
2 |
|
|
|
|
|
36*
|
|
Culture |
ChromeID C. difficile
|
bioMérieux, Marcy-l'Étoile, France |
C. difficile
|
|
|
24 |
|
CDSA |
Becton Dickinson, Sparks, MD, USA |
C. difficile
|
|
|
9 |
|
Blood agar |
|
C. difficile
|
|
|
1 |
|
Home-made |
|
C. difficile
|
|
|
5 |
|
|
|
|
Total |
37†
|
|
GDH |
VIDAS GDH |
bioMérieux, Marcy-l'Étoile, France |
GDH |
Automated EIA |
|
1 |
|
|
|
|
Total |
1 |

Table 2
Combinations of assays types for diagnosis of Clostridium difficile infection according to hospital size
|
Assay type |
Hospital beds (N) |
Total (%) |
|
Toxin AB EIA |
NAAT |
Culture |
GDH |
< 300 |
300–500 |
> 500–1,000 |
> 1,000 |
CL*
|
|
+ |
+ |
+ |
|
1 |
|
14 |
6 |
2 |
23 (40.4) |
|
+ |
+ |
|
|
|
1 |
7 |
1 |
|
9 (15.8) |
|
+ |
|
+ |
|
|
|
5 |
2 |
2 |
9 (15.8) |
|
+ |
|
|
|
1 |
|
6 |
1 |
1 |
9 (15.8) |
|
+ |
+ |
|
1 |
|
2 |
1 |
|
4 (7.0) |
|
+ |
|
|
+ |
|
|
1 |
|
|
1 (1.8) |
|
+ |
|
|
|
|
1 |
|
|
1 (1.8) |
|
|
+ |
|
|
|
1 |
|
|
1 (1.8) |
|
Total |
|
|
|
3 |
1 |
37 |
11 |
5 |
57 (100) |

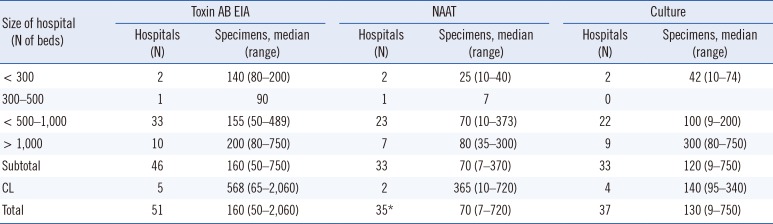
Table 3 shows the median (range) of examined specimens in one month for the toxin AB EIA, NAATs, and
C. difficile culture. More specimens were examined with the toxin AB EIA than with NAATs (
P=0.021). In addition, although the same number of laboratories reported performing NAATs and
C. difficile culture (N=37), there were more specimens examined with the latter method, though this difference was not significant. Moreover, the number of specimens examined using
C. difficile culture was higher for hospitals with ≥1,000 beds than those with 500–1,000 beds (
P=0.008). The number of examined specimens might reflect the disease burden of CDI in the hospital and/or the infection control policy, including the screening strategy for CDI, number of laboratory personnel, and reimbursement of medical insurance. The assays covered by medical insurance were performed more frequently.
Table 3
Numbers of specimens examined for CDI diagnosis according to assay types and hospital size per month in 2015
|
Size of hospital (N of beds) |
Toxin AB EIA |
NAAT |
Culture |
|
Hospitals (N) |
Specimens, median (range) |
Hospitals (N) |
Specimens, median (range) |
Hospitals (N) |
Specimens, median (range) |
|
< 300 |
2 |
140 (80–200) |
2 |
25 (10–40) |
2 |
42 (10–74) |
|
300–500 |
1 |
90 |
1 |
7 |
0 |
|
|
< 500–1,000 |
33 |
155 (50–489) |
23 |
70 (10–373) |
22 |
100 (9–200) |
|
> 1,000 |
10 |
200 (80–750) |
7 |
80 (35–300) |
9 |
300 (80–750) |
|
Subtotal |
46 |
160 (50–750) |
33 |
70 (7–370) |
33 |
120 (9–750) |
|
CL |
5 |
568 (65–2,060) |
2 |
365 (10–720) |
4 |
140 (95–340) |
|
Total |
51 |
160 (50–2,060) |
35*
|
70 (7–720) |
37 |
130 (9–750) |

Toxin AB EIA is more frequently used possibly because of its advantages of short turnaround time and cost-efficiency. However, this assay is often criticized for its poor sensitivity and should therefore no longer be considered as a stand-alone assay for the diagnosis of CDI [
12679101112]. Therefore, the nine (15.8%) laboratories that use only the toxin AB EIA for CDI diagnosis should reconsider their diagnostic strategy.
Since the clinical guidelines for CDI provided by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) were updated in 2010 [
13], many hospitals in the United States have switched the toxin AB EIA to NAATs for CDI diagnosis. Wong et al. [
10] reported that 84.5% of the hospitals surveyed in Ohio, USA, used NAATs as a stand-alone assay in 2014. However, the proportion of laboratories using NAATs as a stand-alone assay was lower in other countries: only 3% and 6% of small (<500 beds) and large (>500 beds) hospitals in Italy in 2012–2013, respectively [
11], 0.9% of participating laboratories in Spain in 2013 [
12], and 11.1% (2/18) of hospitals in Israel in 2012 [
14]. In general, NAATs are more commonly used in combination with other assays, as observed in 16% and 34% of small and large hospitals in Italy in 2012–2013, respectively [
11], 38.2% of participating laboratories in Spain in 2013 [
12], and 38.9% (7/18) of hospitals in Israel in 2012 [
14]. In our study, 36 of 57 (63.2%) of the laboratories conducting CDI assays also used NAATs in combination with other assays, except for one laboratory (1.8%) that reported using NAATs as a stand-alone assay.
Approximately 60% (34/57) of the laboratories reported performing
C. difficile culture, and the majority used chromogenic media for culture (
Table 2), which has been reported to be more sensitive than conventional culture media [
151617]. The
C. difficile cultures performed in many laboratories are not TC, and thus, an additional toxin assay might be needed because not all
C. difficile strains produce toxins [
8]. The extent to which
C. difficile culture is used differs by region: for example, in a 2006 study, only six of the 25 (24%) participating laboratories in Ireland performed
C. difficile culture [
18], whereas 19 of 24 (79%) Finnish laboratories performed it [
19]. In 2012–2013, 25% (38/151) and 37% (24/65) of small and large hospitals in Italy performed
C. difficile culture, respectively, either alone or in combination with other assays [
11]. Given the gap in time between these aforementioned studies, the extent to which NAATs and
C. difficile culture are used for laboratory diagnosis of CDI varies noticeably across countries. However, in our study, more than 80% of the participating hospitals (excluding CL) used NAATs and/or
C. difficile culture with or without the toxin AB EIA. This finding may reflect the greater concern about CDI in Korean hospitals in recent years, which has resulted in the need for more rapid and sensitive diagnosis.
The GDH assay was only recently introduced in the last five years and approved for reimbursement in Korea since 2016. Thus, this assay was not popular at the time of conducting the survey, with only one laboratory reporting its use in combination with the toxin AB EIA. GDH has been reported as a sensitive marker for the detection of
C. difficile and is recommended as a screening assay for CDI diagnosis [
67]; however, GDH-positive results should be followed by an assay to confirm toxin production [
1220].
The recently updated clinical guidelines for CDI by IDSA and SHEA recommend using a stool toxin assay as part of a multistep algorithm (i.e., GDH plus toxin; GDH plus toxin, arbitrated by NAAT; or NAAT plus toxin) rather than an NAAT alone for all specimens received in the clinical laboratory when there are no pre-agreed institutional criteria for patient stool submission. When there are pre-agreed institutional criteria for patient stool submission, it is recommended to use an NAAT alone or a multistep algorithm for testing [
6]. The European Society of Clinical Microbiology and Infectious Diseases (ESCMID) strongly recommends using a two-step algorithm instead of a single assay as a stand-alone assay. The algorithm should start with either the NAAT or GDH assay, and specimens with a positive first assay result should be tested further with the toxin AB EIA. An alternative algorithm is to screen specimens with both the GDH assay and toxin AB EIA [
7].
Although approximately 80% of the laboratories in our study used more than one assay, we did not enquire about the sequences and/or detailed processes used for multiple assays. The diagnostic algorithms applied in Korean hospitals or laboratories are currently not clear; thus, further investigation is necessary to clarify this aspect.
In a survey conducted in Europe in 2014, 24 of the 35 responding countries reported one or more changes in the national/subnational laboratory diagnostics for CDI since 2011 [
9]. The main changes included the availability of commercial diagnostic assays, new or revised guidelines for CDI diagnostics, relevant legislation, and reimbursement policies for diagnostic assays. The main barriers to applying appropriate assays according to the guidelines were financial restrictions, along with insufficient reimbursement and trained staff [
9]. Although this was not explicitly explored in our survey, a similar situation is expected to be occurring in Korea.
There were several limitations in this study. The number and area of participating laboratories were restricted. As mentioned above, the sequences and/or detailed processes used for multiple assays were not investigated, which are the important issues that need to be addressed in order to develop multistep algorithmic approaches for diagnosis of CDI in Korea.
Despite these limitations, this study represents the first survey on the laboratory diagnosis for CDI conducted in Korea. We found considerable variation in the assays used for CDI diagnosis among laboratories in Korea, and some laboratories were still using inappropriate methods such as the toxin AB EIA as a stand-alone assay. NAATs were more rapidly introduced than expected, utilized in approximately 65% of participating laboratories. These findings suggest the need for establishing optimized guidelines for CDI diagnosis in Korea. Thus, our study can provide valuable basic data on the current situation, as a first step towards standardizing laboratory diagnosis of CDI in Korea.







 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download