Abstract
Background
Methods
Results
Acknowledgements
References
Fig. 1
Correlation of time to detection (TTD) between the Virtuo and 3D systems in (A) FA Plus bottles, (B) FN Plus bottles, and (C) total bottles. Total isolates (gram-negative, gram-positive, and yeast) concurrently growing in the Virtuo and 3D systems were compared for TTD. The shaded area indicates the 95% confidence interval of the linear regression line. There were two outlier isolates for which the difference in TTD was more than 20 hours: Klebsiella pneumoniae (detected earlier in the 3D system) and Streptococcus species (detected earlier in the Virtuo system).
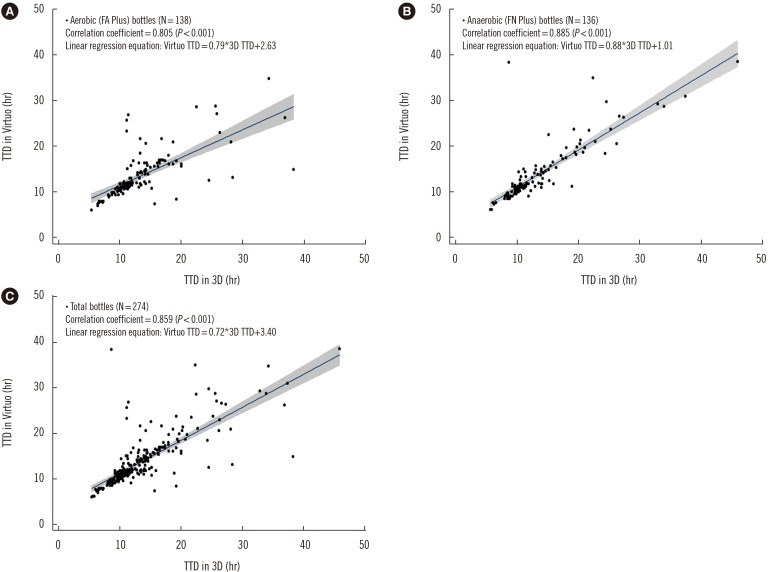
Fig. 2
Cumulative proportions for positives according to time to detection (TTD), showing close similarity between the Virtuo and 3D systems (274 isolates). Approximately 50% of the samples were positive by 12 hours, 80% by 16 hours, and 90% by 24 hours.
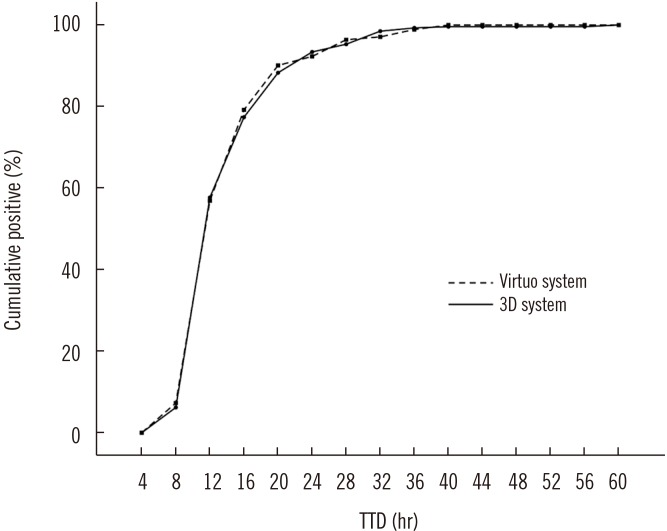
Table 1
Time to detection (hours) of aerobic and anaerobic bottles in the Virtuo and 3D systems
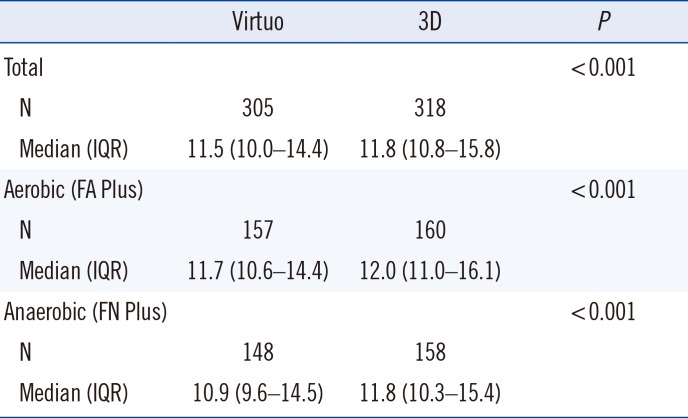
Table 2
Time to detection (hours) of clinically significant monomicrobial pathogens in the Virtuo and 3D systems
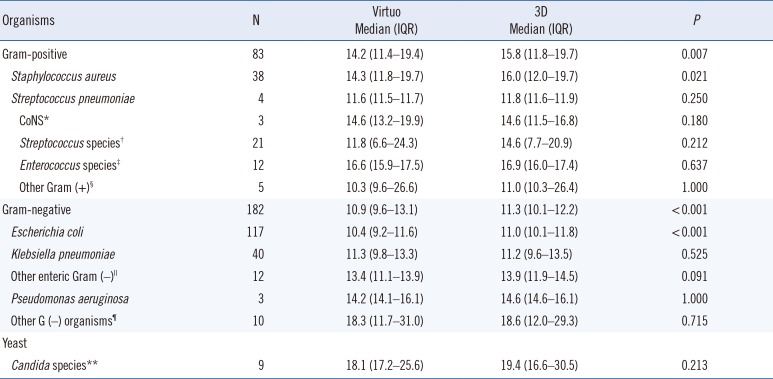
*Includes three Staphylococcus epidermidis; †Includes six Streptococcus anginosus, five Streptococcus dysgalactiae subspecies equisimilis, five Streptococcus agalactiae, two Granulicatella adiacens, one Streptococcus gordonii, one Streptococcus mitis/oralis, and one Streptococcus sanguinis; ‡Includes four Enterococcus faecalis, four Enterococcus faecium, and four Enterococcus hirae; §Includes three Pediococcus acidilactici and two Parvimonas micra; ∥Includes five Enterobacter cloacae, four Enterobacter asburiae, and three Klebsiella oxytoca; ¶Includes three Acinetobacter beijerinckii, two Bacteroides fragilis, two Fusobacterium necrophorum, one Acinetobacter baumannii, one Acinetobacter junii, and one unidentified anaerobic Gram (−) rod; **Includes five Candida tropicalis, two Candida albicans, and two Candida parapsilosis.
Abbreviations: IQR, interquartile range; CoNS, coagulase-negative staphylococci.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download