Abstract
Human cytomegalovirus (HCMV) is a ubiquitous human pathogen and contains double stranded DNA genome with approximately 230 kbp. Molecular genomic studies of HCMV have been attempted in order to understand the pathogenesis and evolution of HCMV. However, studies on HCMV strains of Asian origin are limited. In this study, it was attempted to understand the genomics of HCMV isolated from Korea. Clinical strain LCW isolated from Korean patient was passaged in vitro cell culture, and subjected to next-generation sequencing. Complete genome sequence was obtained and compared with other HCMV strains. The LCW genome was found to contain 170 open reading frames (ORFs) and two ORF (RL5A and RL13) of the strain LCW were found to be truncated due to early stop codon. Phylogenetic analysis suggested that the strain LCW was closely related with Asian strains such as HCMV strains JHC and HAN. Common nucleotide sequences among the 3 Asian strains distinguishable from other strains were detected at 197 sites including 104 sites in ORFs.
Figures and Tables
 | Figure 1Genome map of the HCMV strain LCW. Nucleotide positions corresponding to the HCMV genome structure and the ORFs contained in each of the genome structure is indicated. |
 | Figure 2Sequence alignment of HCMV ORF RL5A and RL13. Early stops due to mutations in the strain LCW are boxed. (A) ORF RL5A due to T->A substitution at the third position of the codon, (B) RL13 due to C->A substitution at the first position of the codon. |
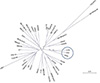 | Figure 3Phylogenetic tree of 42 HCMV strains. Genomic DNA sequences of HCMV strains were retrieved from NCBI GenBank database and aligned with ClustalW program together with the genome sequence of the Korean strain LCW. Phylogenetic tree was constructed using neighbor-joining model in Phylip package. Cluster of HCMV strains of Asian origin is indicated as a circle. |
References
1. Sinclair J, Reeves M. The intimate relationship between human cytomegalovirus and the dendritic cell lineage. Front Microbiol. 2014; 5:389.

2. Weller TH. The cytomegaloviruses: ubiquitous agents with protean clinical manifestations. N Engl J Med. 1971; 285:203–214.

3. Landolfo S, Gariglio M, Gribaudo G, Lembo D. The human cytomegalovirus. Pharmacol Ther. 2003; 98:269–297.

4. Hassan J, Connell J. Translational Mini-Review Series on Infectious Disease: Congenital cytomegalovirus infection: 50 years on. Clin Exp Immunol. 2007; 149:205–210.

5. Boeckh M, Geballe AP. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest. 2011; 121:1673–1680.

6. Staras SA, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin Infect Dis. 2006; 43:1143–1151.

7. Jesionek A, Kiolemenoglou B. Ueber einen befund von protozoenartigen gebilden in den organen eines hereditar-luetis-chen foetus. Muenchner Med Wochenschr. 1904; 51:1905–1907.
8. Smith MG. Propagation in tissue cultures of a cytopathogenic virus from human salivary gland virus (SGV) disease. Proc Soc Exp Biol Med. 1956; 92:424–430.

9. Craig JM, Macauley JC, Weller TH, Wirth P. Isolation of intranuclear inclusion producing agents from infants with illnesses resembling cytomegalic inclusion disease. Proc Soc Exp Biol Med. 1957; 94:4–12.

10. Elek SD, Stern H. Development of a vaccine against mental retardation caused by cytomegalovirus infection in utero. Lancet. 1974; 1:1–5.

11. Plotkin SA, Furukawa T, Zygraich , Huygelen C. Candidate cytomegalovirus strain for human vaccination. Infect Immun. 1975; 12:521–527.

12. Just M, Buergin-Wolff A, Emoedi G, Hernandez R. Immunisation trials with live attenuated cytomegalovirus TOWNE 125. Infection. 1975; 3:111–114.

13. Brayman KL, Dafoe DC, Smythe WR, Barker CF, Perloff LJ, Naji A, et al. Prophylaxis of serious cytomegalovirus infection in renal transplant candidates using live human cytomegalovirus vaccine. Interim results of a randomized controlled trial. Arch Surg. 1988; 123:1502–1508.

14. Khanna R, Diamond DJ. Human cytomegalovirus vaccine: time to look for alternative options. Trends Mol Med. 2006; 12:26–33.

15. Krause PR, Bialek SR, Boppana SB, Griffiths PD, Laughlin CA, Ljungman P, et al. Priorities for CMV vaccine development. Vaccine. 2013; 32:4–10.

16. Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990; 154:125–169.

17. Dargan DJ, Douglas E, Cunningham C, Jamieson F, Stanton RJ, Baluchova K, et al. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J Gen Virol. 2010; 91:1535–1546.

18. Renzette N, Gibson L, Bhattacharjee B, Fisher D, Schleiss MR, Jensen JD, et al. Rapid intrahost evolution of human cytomegalovirus is shaped by demography and positive selection. PLoS Genet. 2013; 9:e1003735.

19. Fernández-Ruiz M, Corrales I, Arias M, Campistol JM, Giménez E, Crespo J, et al. Association between individual and combined SNPs in genes related to innate immunity and incidence of CMV infection in seropositive kidney transplant recipients. Am J Transplant. 2015; 15:1323–1335.

20. Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A. 2003; 100:14976–14981.

21. Novak Z, Ross SA, Patro RK, Pati SK, Kumbla RA, Brice S, et al. Cytomegalovirus strain diversity in seropositive women. J Clin Microbiol. 2008; 46:882–886.

22. Renzette N, Pokalyuk C, Gibson L, Bhattacharjee B, Schleiss MR, Hamprecht K, et al. Limits and patterns of cytomegalovirus genomic diversity in humans. Proc Natl Acad Sci U S A. 2015; 112:E4120–E4128.

23. Lee GC, Lee DG, Choi SM, Yoo JH, Park SH, Choi JH, et al. Use of Time-Saving Flow Cytometry for Rapid Determination of Resistance of Human Cytomegalovirus to Ganciclovir. J Clin Microbiol. 2005; 43:5003–5008.

24. Jung GS, Kim YY, Kim JI, Ji GY, Jeon JS, Yoon HW, et al. Full genome sequencing and analysis of human cytomegalovirus strain JHC isolated from a Korean patient. Virus Res. 2011; 156:113–120.

25. Davison AJ, Akter P, Cunningham C, Dolan A, Addison C, Dargan DJ, et al. Homology between the human cytomegalovirus RL11 gene family and human adenovirus E3 genes. J Gen Virol. 2003; 84:657–663.

26. Cunningham C, Gatherer D, Hilfrich B, Baluchova K, Dargan DJ, Thomson M, et al. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J Gen Virol. 2010; 91:605–615.

27. Wilkinson GW, Davison AJ, Tomasec P, Fielding CA, Aicheler R, Murrell I, et al. Human cytomegalovirus: taking the strain. Med Microbiol Immunol. 2015; 204:273–284.

28. Sijmons S, Thys K, Mbong Ngwese M, Van Damme E, Dvorak J, Van Loock M, et al. High-throughput analysis of human cytomegalovirus genome diversity highlights the widespread occurrence of gene-disrupting mutations and pervasive recombination. J Virol. 2015; 89:7673–7695.





 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download