Abstract
Laboratory inbred mice are used widely and commonly in biomedical research, but inbred mice do not have a big enough gene pool for the research. In this study, genetic and morphometric analyses were performed to obtain data on the characteristics of a newly developing inbred strain (KWM/Hym) captured from Chuncheon, Korea. All of five Korean wild male mice have the zinc-finger Y (ZfY) gene. Also, all of 19 Korean wild mice used in this analysis have the AKV-type murine leukemia virus gene, indicating that Korean wild mice might be Mus musculus musculus. To identify the genetic polymorphism in KWM/Hym, SNP analysis was performed. In a comparison with 28 SNP markers, there was a considerable difference between KWM/Hym and several inbred strains. The homogeneity between KWM/Hym and the inbred strains was as follows: C57BL/6J (39.3%), BALB/c AJic (42.9%), and DBA/2J (50%). KWM/Hym is most similar to the PWK/PhJ inbred strain (96.4%) derived from wild mice (Czech Republic). To identify the morphometric characteristics of KWM/Hym, the external morphology was measured. The tail ratio of male and female was 79.60±3.09 and 73.55±6.14%, respectively. KWM/Hym has short and agouti-colored hairs and its belly is white with golden hair. Taking these results together, KWM/Hym, a newly developing inbred mouse originated from wild mouse, might be use as new genetic resources to overcome the limitations of the current laboratory mice.
House mice (Mus musculus) are the most used and primary model organism for human diseases such as metabolic, aging, cancer, and immunological disorders. Mice and humans are similar physiologically as well as genetically [1]. Mouse is small, easy to reproduce all year, easily restricted to genetic and biological variables, cheap to feed, and has strong tolerance to inbreeding compared with other mammal species [2]; however, on the other hand, the uniform inbred mice lack genetic diversity because they originated from a relatively limited genetic pool [3]. Numerous efforts have been made to develop the “next-generation mice” to overcome the limitations of inbred mice models. As part of these efforts, the “MODEL-AD consortium” was launched by the National Institutes of Health (NIH) in 2016 to develop and supply model animal mice that had overcome such limitations and are more suitable for experimental purposes [4]. Thus, the development of a new inbred mouse derived from wild mouse can be a new genetic variation in the current mouse resources.
The geological origin of the Mus genus musculus species is India. House mice are classified into 11 subspecies by their morphological characters and geographic distribution [5]. It is suggested that house mice might be grouped into four subspecies (domesticus, bactrianus, castaneus, and musculus) based on their chromosome C-band pattern [6], biochemical markers [7], mitochondria DNA [8], ribosomal DNA [9], and other genetic characters [10]. Inbred strains of laboratory mice showed a mosaic genome derived in a polygenic manner from M. m. domesticus, M. m. musculus, and M. m. castaneous according to genotypic analysis, and a cross bred species M. m. molossinus between M. m. musculus and M. m. castastaneous was reported from Japan. However, nowadays, most commonly used inbred mice are M. m. domesticus [3]. It is reported that Korean wild mice have characteristics of both M. m. musculus and M. m. molossinus [11]. But their genetic and phenotypic features are not fully understood.
Zinc-finger Y (ZfY) genes located on the Y chromosome have two type, Zfy-1 and Zfy-2. These ZfY genes can be used in characterizing the subspecies by the presence (in musculus and castaneus), or absence (in domesticus) [12]. AKV-type murine leukemia virus (MuLV) gene was found in Korean mice, Japanese mice (M. m. molossinus), and mainly northern Chinese mice. Thus, ZfY and AKV-type MuLVs genes might be useful tools as specific genetic markers for distinguishing subspecies in Mus musculus.
Single nucleotide polymorphisms (SNPs) are the most abundant class of genetic markers in the genome. Since most SNPs have only common alleles, genotyping methods are comparatively easy. Genetic monitoring was usually has been used for detecting genetic contamination in the inbred mouse based on assaying single-nucleotide polymorphism (SNP) markers positioned on all autosomes and the X chromosome [13]. SNP analysis also was performed to identify genetic polymorphism differences between laboratory mice and wild mice. It is reported that methods for genetic monitoring in mice has used physical characteristics such as immunological (major histocompatibility complex differences) and biochemical methods (isoenzymes) as well as coat color [14]. Morphometrical characteristics such as body parameters (size, weight, length, tail ratio, skull length, etc.) and coat color are highly variable for different mouse strains depending on their genetic and environmental backgrounds [15]. Wild mice have short hair that is colored from light to dark agouti (or brown), but the coat of laboratory mice is produced in a lot of colors from white to black. Some subspecies of Mus musculus have a light-colored belly (white to cream). The ears and tail have little hair [16].
In this study, genetic and morphometric analyses were carried out to obtain data on the characteristics of wild mouse captured from Chuncheon, Korea. The results suggest that this newly developing inbred mouse (KWM/Hym) originated from wild mouse might be useful for genetic research and may become a valuable tool for overcoming the limitations of current inbred mice.
The wild mice were captured between March and April of 2017 and 2018 in Chuncheon-si, Gangwon-do, Korea. The captured mice were kept in a conventional animal care facility that maintained a regular environment: 22±2℃, 55±10% relative humidity, and a 12 hr light and 12 hr night routine cycle. Sweet potato and normal rodent pellet feeds (Cargill Agri Purina, Seongnam, Gyeonggi, Korea) and water were provided ad libitum. The animal study was conducted in accordance with the regulations of the Institutional Animal Care and Use Committees of Hallym University (Hallym-2017-5).
Tail genomic DNA was used for subspecies and genetic screening. Tail tips no longer than 1–2 mm were incubated with a buffer (1X SSC: 439 µL, 1 M Tris-HCl: 5 µL, 0.5 M EDTA: 1 µL, 10% SDS: 50 µL, 20 mg/mL Proteinase K (Sigma): 5 µL) at 53℃ for 5 hr. Then the genomic DNA was purified by phenol, phenol/chloroform, chloroform, and isopropanol reagents. After that, genomic DNA was washed with 70% ethanol and 100% ethanol. For polymerase chain reaction (PCR), the genomic DNA was dissolved in 50 µL of Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA).
Amplification of ZfY and AKV-type MuLV markers was performed using the procedures of Suh et al. with minor modifications [17]. Primer sequences were as follows: ZfY, 5′CATTAAGACAGAAAAGACCACCG-3′ and 5′GTGAGGAAATTTCTTCCTGTGG-3′; AKV-type MuLV, 5′GATCTCTGTATGTTGGCCCTCCA-3′ and 5′GGTCATGTCCAGAGACGTATAGG-3′. PCR was carried out in PCR reaction buffer containing rTaq Plus 5× PCR Master Mix reagent (ELPiS, Daejeon, Korea), 50 ng genomic DNA, and 20 p mole of each primer. PCR condition was subjected to initial denaturation at 95℃ for 3 min followed by 33 cycles of 95℃ for 60 sec, 59℃ for 90 sec, and 72℃ for 90 sec and then a final elongation period at 72℃ for 2 min. The amplified PCR products were separated by electrophoresis on 3% agarose gel. Gels were subsequently stained with ethidium bromide and photographed under UV light. The PCR product band sizes of ZfY and AKV-type MuLV markers were 202 bp (and 184 bp) and 530 bp, respectively.
SNP analysis for genetic monitoring was performed using the procedures of Petkov et al. with minor modifications [13]. Publicly available SNPs were selected from the Roche database (mouseSNP.roche.com) based on their chromosomal positions and strain distribution information. SNP determination was performed by real-time PCR (7900HT Fast System, The Applied Biosystems, ThermoFisher Scientific, USA) using fluorescent-based PCR. PCR condition was subjected to initial denaturation at 95℃ for 10 min followed by 45 cycles of 95℃ for 15 sec, 92℃ for 15 sec, and 60℃ for 1 min. The sequence detection was performed using Systems Software version 2.3 (The Applied Biosystems).
Total body length, tail length, ear length, head length, hind foot length, body weight, and tail ratio were measured according to Marshall's method [18], and statistical analysis was performed. Data are presented as the mean±standard deviation (SD). Hairs were gently plucked from the back and belly of mice under inhalation anesthesia. Then the pigment pattern of the hairs was observed under an Axioscope microscope (Carl Zeiss, Germany).
The ZfY gene is identified in Korea wild mouse by bands of 202 and 184 bp (Figure 1). Because the ZfY gene is actively transcribed in males, all of Korean wild mice have bands of 202 and 184 bp (Figure 1). All of 19 Korea wild mice have an AKV-type MuLV gene (530 bp). Therefore, the Korean wild mouse is considered to be similar to M. m. musculus.
Genetic monitoring in developing inbred mice (generation one to four) derived from wild mice was performed using SNP markers. When comparing KWM/Hym with several inbred strains, it was confirmed to have the following homogeneity: C57BL/6J (39.3% homogeneity, 11 match markers), BALB/c AJic (42.9% homogeneity, 12 match markers), and DBA/2J (50% homogeneity, 14 match markers) (Table 1). The results showed that there was a considerable difference between KWM/Hym and laboratory inbred strains and also that KWM/Hym is most similar to the PWK/PhJ inbred strain derived from wild mice (Czech Republic). The homogeneity between KWM/Hym and the PWK/PhJ strain is 96.4%, only the re3022977 marker out of 28 SNP markers was different from C to T (Table1). However, in this study, polymorphism within KWM/Hym was not observed.
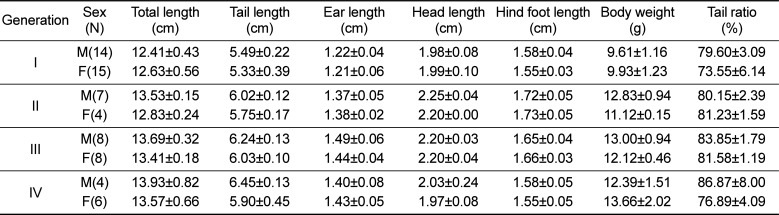
External morphology such as total body length, tail length, ear length, head length, hind foot length, tail ratio, and body weight were analyzed to identify the morphometric characteristics of Korean wild mice and KWM/Hym. Twenty-nine Korean wild mice (14 males/15 females) were used for the analysis. In males, total body length and tail ratio were 12.41±0.43 mm and 79.60±3.09%, respectively. In females, total body length and tail ratio were 11.76±0.97 mm and 81.13±8.00%, respectively. No significant differences between the male and female were found in any of the parameters (Table 2). Tail ratio in both sexes was not significantly different with in-breeding. Body weight increased slightly in accordance with in-breeding (Table 2).
The Korean wild mouse has short and flexible fur. Its head and back are agouti-colored and its belly is white with golden hair (Figure 2A). The hair color of a mouse is usually determined by the color of its upper hair. The upper-hair is divided into monotrich, awl, and auchen, and the lower-hair consists of zigzag hair (Figure 2B).
Recently, a lot of attempts have been made to overcome the limitations of current laboratory mice. One of the efforts is the production of a transgenic mouse, which is useful but expensive and time-consuming. Another attempt is the development of a new inbred strain derived from wild mouse. But there are also many basic biological issues such as parasite infection, disease as well as a lack of clarification for genetic and phenotypic variations. Therefore, in this study, we identified the genetic and morphological characteristics of Korean wild mice captured at Chuncheon, Korea.
To identify the subspecies of Korean wild mice, the distribution of specific subspecies markers (ZfY and AKV-type MuLV) were analyzed. The ZfY gene located on the Y chromosome is actively transcribed in males and is identified in Korean wild mice (100%, all five males) by bands of 202 and 184 bp (Figure 1). AKV-type MuLV gene is identified all of 19 Korea wild mice (Figure 1). The geographic origins of Mus musculus subspecies is India, and from there it spread all over the world by following human migration routes [19]. Mus musculus is often classified into three principal subspecies: M. m. domesticus, M. m. musculus, and M. m. castaneous. M. m. domesticus is indigenous to western Europe, but it has been transported around the world to areas such as Africa, the Near East, the Americas, and Australia by human migrants. M. m. musculus is found from northern and eastern Europe across to Asia, and M. m. castaneous occupies areas in Southeast Asia [20]. It is reported that M. m. molossinus arose from a hybrid of M. m. musculus and M. m. castaneus in Japan. The Korean wild mouse is considered to be similar to M. m. musculus.
For most of the twentieth century, research on mice was dominated by laboratory inbred strains. But it became apparent from research of inbred strains captured from wild mice that there were lots of numbers of naturally occurring polymorphisms rather than laboratory inbred mice, which are homogeneous over 98.6% of their genomes [21]. Genetic monitoring of KWM/Hym was performed using SNP markers. In a comparison of KWM/Hym with several inbred strains, it was confirmed to have the following homogeneity: C57BL/6J (39.3%), BALB/c AJic (42.9%), and DBA/2J (50%); therefore, there was a considerable difference between KWM/Hym and laboratory inbred strains (Table 1). The results showed that KWM/Hym is most similar to the PWK/PhJ inbred strain derived from wild mice (Czech Republic). The homogeneity between KWM/Hym and PWK/PhJ strain is 96.4%, and only the re3022977 marker out of 28 SNP markers was different from C to T [13]. SKIVE (Denmark), CZECHI/EiJ, CZECHII/EiJ, and PWK/PhJ (Czech Republic) are inbred strains derived from wild mice of M. m. musculus [14]. Thus, KWM/Hym, a newly developing inbred mouse derived from wild mice, is considered to be similar to M. m. musculus.
To identify the morphometric characteristics in Korean wild mice, external morphology such as total body length, tail length, ear length, head length, hind foot length, tail ratio, and body weight were analyzed. The tail ratio of male and female in Korean wild mice are showed 79.60±3.09 and 81.13±8.00%, respectively, indicating that there are no significant differences between male and female (Table 2). In spite of inbreeding, the tail ratio was not changed in developing inbred mice (KWM/Hym). The Korean wild mouse has short and flexible fur. The coat of back and belly are agouti-colored and white with golden hair, respectively (Figure 2A). The hair types are very similar to inbred strains derived from M. m. musculus. Taking these results together, Korean wild mice captured from Chuncheon and KWM/Hym, a newly developing inbred mice, are considered to be similar to M. m. musculus. KWM/Hym might be used as a valuable resource for overcoming the limitations of current laboratory mice.
Acknowledgments
This work was supported by Mouse Phenotyping Project (KMPC:2014M3A9D5A01075129) of the Ministry of Science, ICT and Future Planning through the National Research Foundation and Basic Science Research Program (NRF-2016R1D1A2B02011858).
References
1. Bedell MA, Jenkins NA, Copeland NG. Mouse models of human disease. Part I: techniques and resources for genetic analysis in mice. Genes Dev. 1997; 11(1):1–10. PMID: 9000047.

2. The mouse in biomedical research. 2nd Edition. Academic Press;2006.
3. Abe K, Noguchi H, Tagawa K, Yuzuriha M, Toyoda A, Kojima T, Ezawa K, Saitou N, Hattori M, Sakaki Y, Moriwaki K, Shiroishi T. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res. 2004; 14(12):2439–2447. PMID: 15574823.

4. Reardon S. Frustrated Alzheimer's researchers seek better lab mice. Nature. 2018; 563(7733):611–612. PMID: 30482928.

5. Schwarz E, Schwarz HK. The wild and commensal stocks of the house mouse, Mus musculus. J Mammal. 1943; 24:59–72.
6. Boursot P, Auffray JC, Britton-Davidian J, Bonhomme F. The evolution of house mice. Annu Rev Ecol Syst. 1993; 24:119–152.

7. Bonhomme F, Catalan J, Britton-Davidian J, Chapman VM, Moriwaki K, Nevo E, Thaler L. Biochemical diversity and evolution in the genus Mus. Biochem Genet. 1984; 22(3-4):275–303. PMID: 6375655.

8. Yonekawa H, Moriwaki K, Gotoh O, Hayashi JI, Watanabe J, Miyashita N, Petras ML, Tagashira Y. Evolutionary relationships among five subspecies of Mus musculus based on restriction enzyme cleavage patterns of mitochondrial DNA. Genetics. 1981; 98(4):801–816. PMID: 6277733.

9. Suzuki H, Miyashita N, Moriwaki K, Kominami R, Muramatsu M, Kanehisa T, Bonhomme F, Petras ML, Yu ZC, Lu DY. Evolutionary implication of heterogeneity of the nontranscribed spacer region of ribosomal DNA repeating units in various subspecies of Mus musculus. Mol Biol Evol. 1986; 3(2):126–137. PMID: 2895411.
10. Moriwaki K, Shiroishi T, Yonekawa H. Wild mouse from a geneticist's viewpoint. Japan Scientific Societies Press. Genetics in Wild Mice. Tokyo: Karger;1994. p. 13–25.
11. Lee YH, Park JY, Yun YM, Oh SH, Lee JE, Jin HK, Do SG, Suh JG, Lee CH, Hyun BH, Oh YS, Seong JK. Genetic characteristics of Korean wild mice (Mus musculus spp.) by biochemical marker gene. Lab Anim Res. 2000; 16(1):33–39.
12. Duplantier JM, Orth A, Catalan J, Bonhomme F. Evidence for a mitochondrial lineage originating from the Arabian peninsula in the Madagascar house mouse (Mus musculus). Heredity (Edinb). 2002; 89(2):154–158. PMID: 12136419.

13. Petkov PM, Cassell MA, Sargent EE, Donnelly CJ, Robinson P, Crew V, Asquith S, Haar RV, Wiles MV. Development of a SNP genotyping panel for genetic monitoring of the laboratory mouse. Genomics. 2004; 83(5):902–911. PMID: 15081119.

14. Shao Y, Wang J, Qiao Y, He Y, Cao W. Morphological variability between wild populations and inbred stocks of a Chinese minnow, Gobiocypris rarus. Zoolog Sci. 2007; 24(11):1094–1102. PMID: 18348610.

15. Yom-Tov Y, Geffen E. Geographic variation in body size: the effects of ambient temperature and precipitation. Oecologia. 2006; 148(2):213–218. PMID: 16525785.

16. Berry RJ. Natural history of the house mouse. Field Studies Council;1970. 3:p. 219–262.
17. Hwang SH, Cho KI, Park JH, Ryoo ZY, Oh YS, Suh JG. Studies on distribution of the Zfy, Fv4r, and AKV-type MuLV genes in Korean wild mice. Lab Anim Res. 2005; 21(2):135–139.
18. Marshall JT. Taxonomy in “The mouse in biomedical research”. Academic Press;1981. p. 17–26.
19. Auffray C, Tchernov E, Bonhomme F, Giora Heth S, Simson S, Nevo E. Presence and ecological distribution of Mus “spretoides” and Mus musculus domesticus in Israel-Circum-Mediterranean vicariance in the genus Mus. Z. Saugetierkunde. 1990; 55:1–10.
20. Boursot P, Din W, Anand R, Darviche D, Dod B, Von Deimling F, Talwar GP, Bonhomme F. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J Evol Biol. 1996; 9(4):391–415.

21. Barton NH, Keightley PD. Understanding quantitative genetic variation. Nat Rev Genet. 2002; 3(1):11–21. PMID: 11823787.

Figure 1
Distribution of ZfY and AKV-type MuLV genes in Korean wild mice. Bands of 202 and 184 bp and 530 bp indicate distribution of Zfy and AKV-type MuLV genes, respectively. Mark: 100 bp DNA ladder.
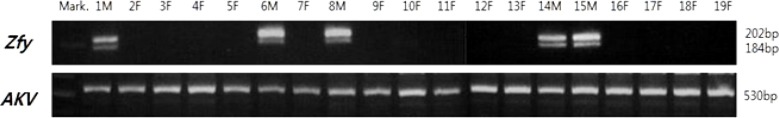
Figure 2
Photograph and coat color of Korean wild mice. (A) Photograph of Korean wild mouse. Korean wild mouse has agouti-colored back and white belly with golden hair. (B) The four main hair types. Monotrich (a), awl (b), auchen (c), and zigzag (d).
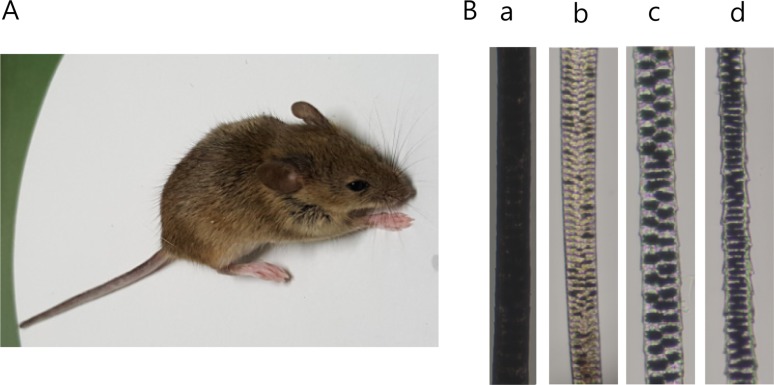
Table 1
Genetic monitoring in KWM/Hym using SNP markers
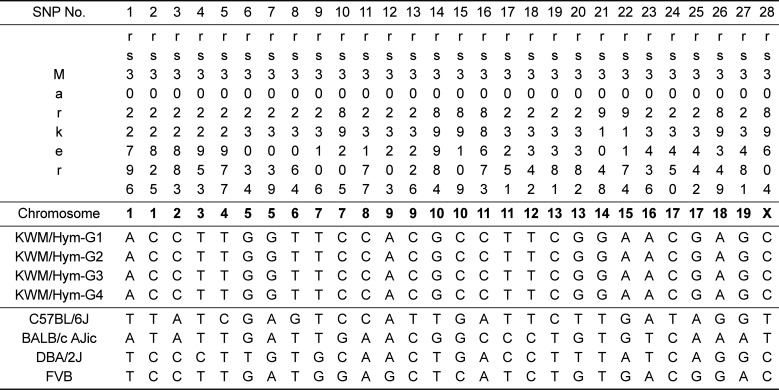
Table 2
Morphometric characteristics of Korean wild mice and KWM/Hym




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download