INTRODUCTION
METHODS
Study population and clinicopathologic data
Immunohistochemistry and molecular tests
Statistical analysis
RESULTS
Patient clinicopathologic characteristics according to PIK3CA mutation status
Table 1
Clinicopathologic parameters according to PIK3CA mutation status
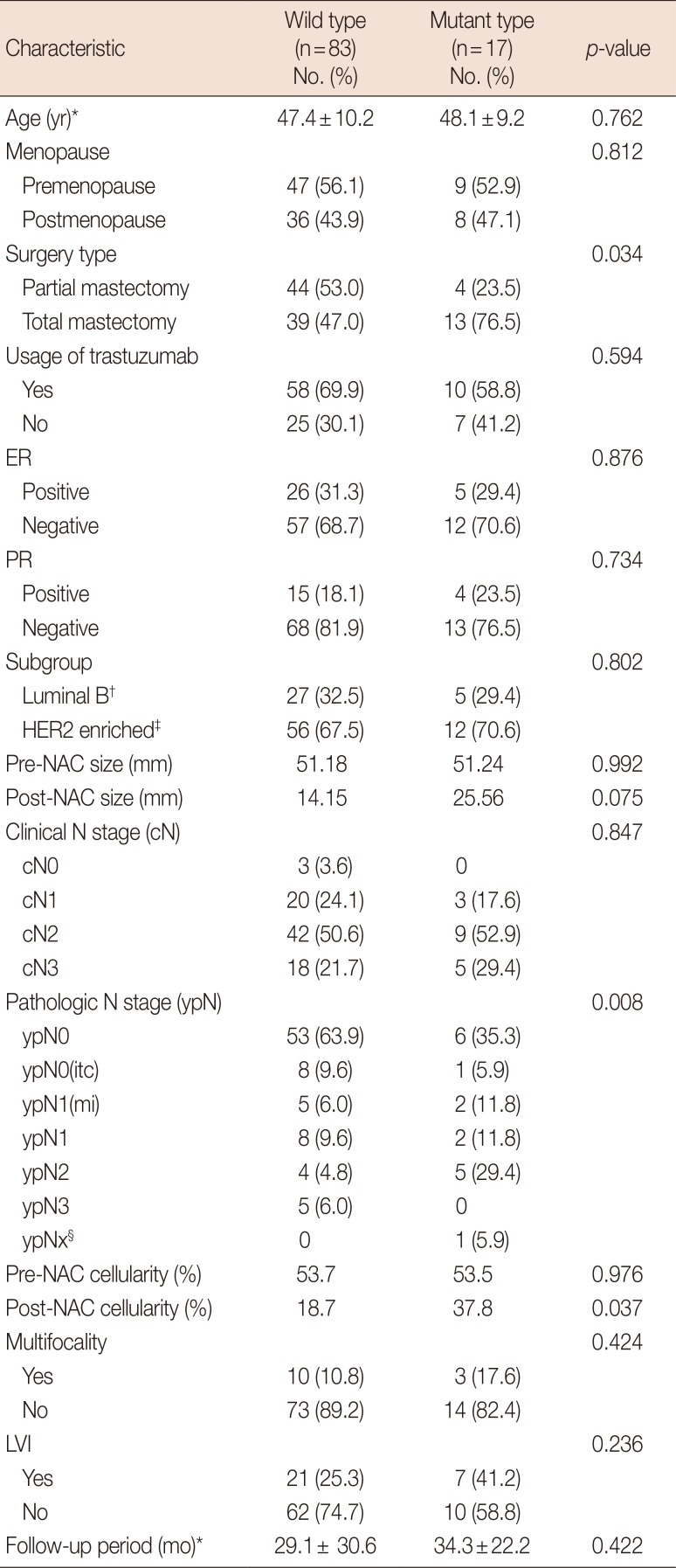
ER=estrogen receptor; PR=progesterone receptor; HER2=human epidermal growth factor receptor 2; NAC=neoadjuvant chemotherapy; itc=isolated tumor cells; mi=microinvasion; LVI=lymphovascular invasion.
*Mean±SD; †Luminal B: immunonistochemical stain for estrogen receptor or progesterone receptor (+); ‡HER2 enriched: immunonistochemical stain for estrogen receptor and progesterone receptor (−); §The one of the ypNx case did not undergo node dissection.
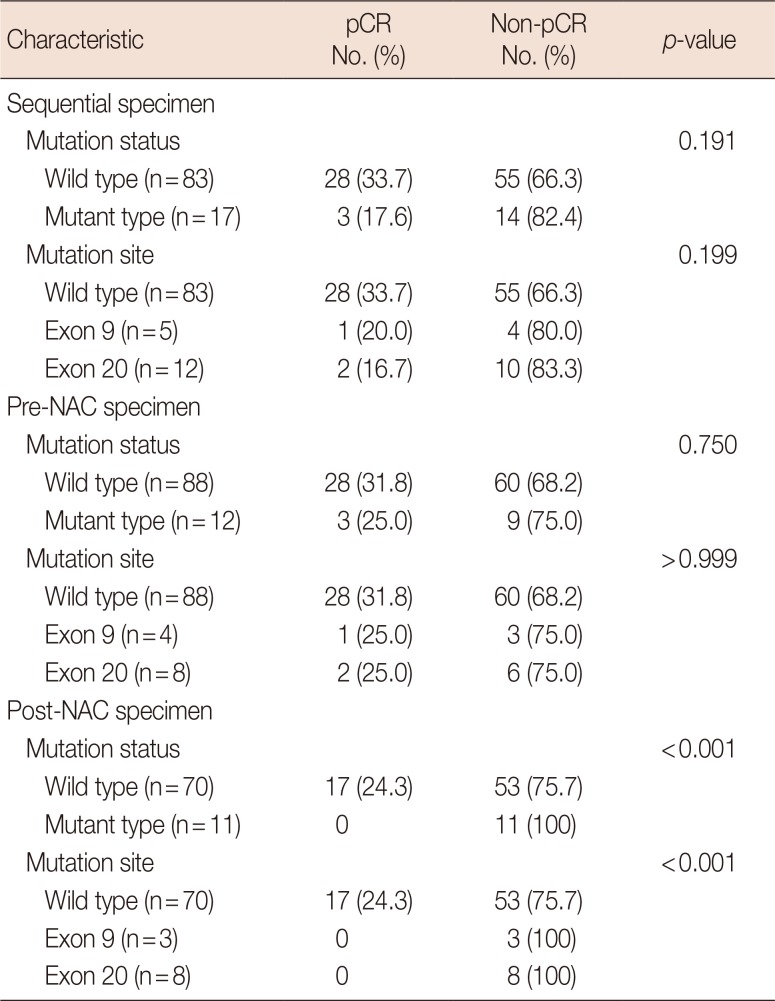
Pathologic response with neoadjuvant chemotherapy and PIK3CA mutation status in sequential specimens
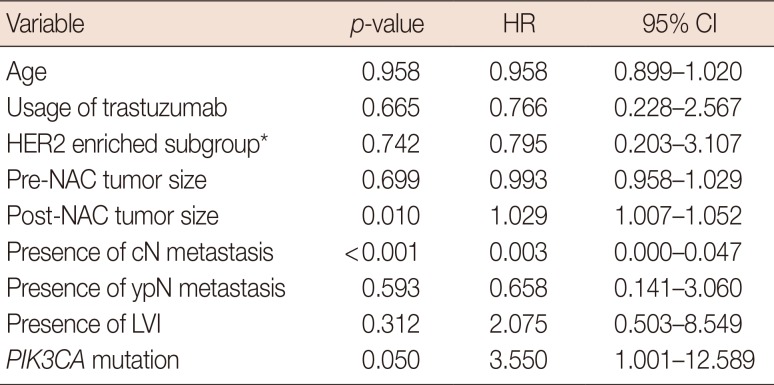
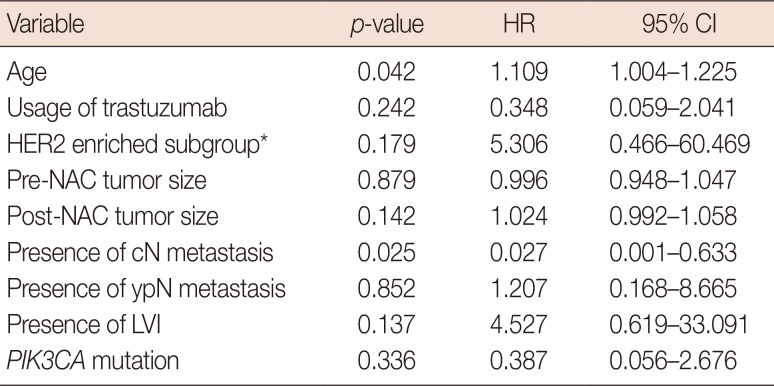
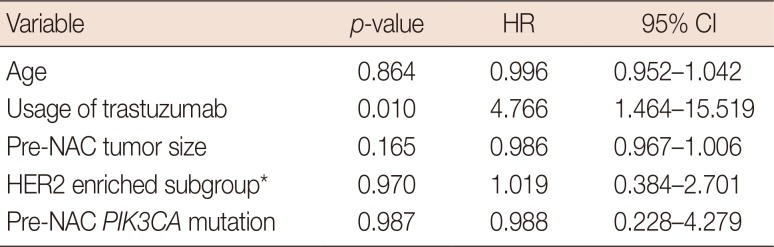
Survival analysis
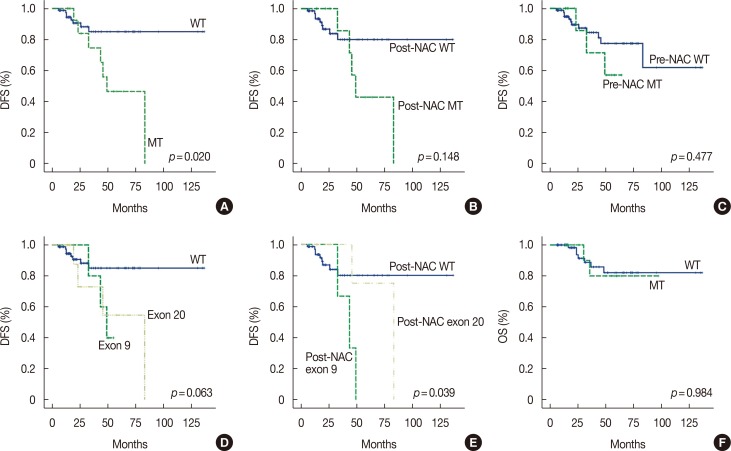
Figure 1
Survival curves for disease-free survival (DFS) and overall survival (OS). (A) PIK3CA mutant group (MT) in any sequential specimen had a shorter median DFS than the purely wild-type group (WT) in sequential specimens (58.3 months vs. 119.3 months, p=0.020). (B) In post-neoadjuvant chemotherapy (NAC) specimens, MT showed a shorter median DFS tendency than WT (60.2 months vs. 113.5 months, p=0.148). (C) Median DFS was not significantly different in pre-NAC WT and pre-NAC MT (51.8 months vs. 103.9 months, p=0.477). (D) There was no difference in median DFS depending on mutation site in sequential analysis (119.3 months vs. 47.2 months vs. 59.8 months, p=0.063). (E) Mutation site was associated with different median DFS in post-NAC specimens (post-NAC WT: 113.5 months vs. post-NAC exon 9: 41.8 months vs. post-NAC exon 20: 74.0 months, p=0.039). In subgroup analysis, post-NAC exon 9 mutation was found to have significantly shorter DFS than other groups (vs. post-NAC WT: p=0.017 vs. post-NAC exon 20: p=0.048). (F) OS was not correlated with PIK3CA mutation status (84.5 months vs. 118.0 months, p=0.984).
Prognostic or predictive effectiveness of PIK3CA mutation
Change of PIK3CA mutation status in sequential specimens
Table 6
Clinicopathologic characteristics of cases with mutation change in sequential analysis
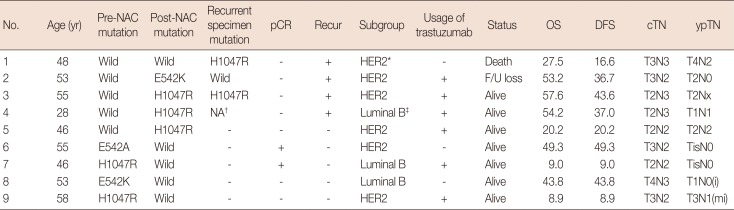
NAC=neoadjuvant chemotherapy; pCR=pathologic complete response; OS=overall survival; DFS=disease-free survival; cTN=clinical T&N stage; ypTN=post-NAC pathological T&N stage; HER2=human epidermal growth factor receptor 2; F/U=follow-up; i=isolated tumor cell; mi=microinvasion.
*HER2 enriched: immunonistochemical stain for estrogen receptor and progesterone receptor (−); †NA: multiple distant metastases were found in the brain in this case, but further tests could not be performed because no samples were obtained; ‡Luminal B: immunonistochemical stain for estrogen receptor or progesterone receptor (+).
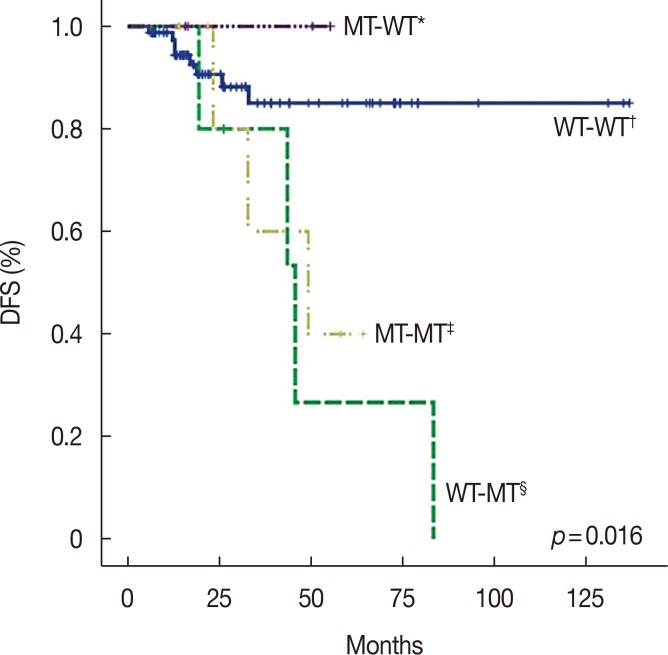
Figure 2
Survival curve for disease-free survival (DFS) according to mutation change status. There are four groups of mutation change status in sequential analysis. Due to lack of recurrence in the mutant-wild (MT-WT) group, Kaplan-Meier methods could not be utilized. The log-rank test was used to reveal significantly different survival curves for DFS (p=0.016). Subgroup analysis determined a significant DFS difference between the WT-WT group and WT-MT groups (p=0.005).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download