Abstract
Purpose
Methods
Results
Conclusion
References
Figure 1
Identification of differentially expressed messenger RNAs (mRNAs) in the The Cancer Genome Atlas (TCGA) BRCA database. (A) Heat map of the log2-fold expression changes in 10 B7 family members mRNAs in the TCGA BRCA database (n=1,092). The horizontal row represents different mRNAs, and the vertical column represents different patients. Green squares indicate increases, and red squares indicate decreases. (B) Statistical analysis of altered expression of B7 family members in the TCGA BRCA data. The false discovery rate for all data is less than 0.01.
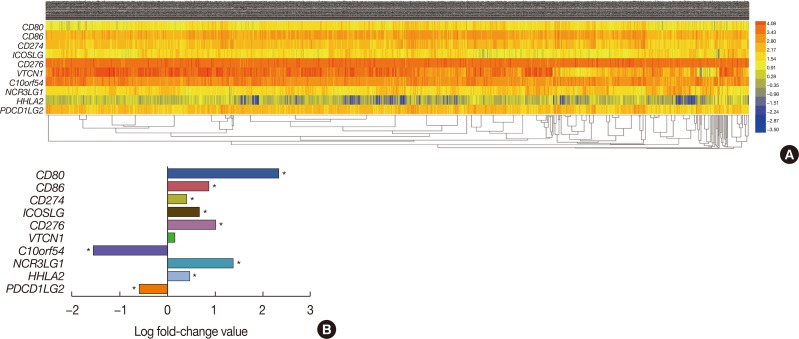
Figure 2
Relationship between six B7 family members and overall survival in breast cancer patients. (A) Using The Cancer Genome Atlas survival data and Cutoff Finder (http://molpath.charite.de/cutoff/), we correlated the messenger RNA (mRNA) expression level of B7 family members with overall survival. Six B7 family members, CD274, CD276, HHLA2, ICOSLG, NCR3LG1, and PDCD1LG2, had prognostic value in patients with breast cancer (log-rank p<0.05). Notably, high CD274, HHLA2, and PDCD1LG2 expression correlated with longer survival, whereas high CD276, ICOSLG, and NCR3LG1 expression correlated with shorter survival. (B) Real-time polymerase chain reaction was performed to determine CD274, CD276, HHLA2, ICOSLG, NCR3LG1, and PDCD1LG2 expression in MCF7, MDA-MB-231, SK-BR-3 and human fibrocystic disease epithelium cell lines MCF10A.
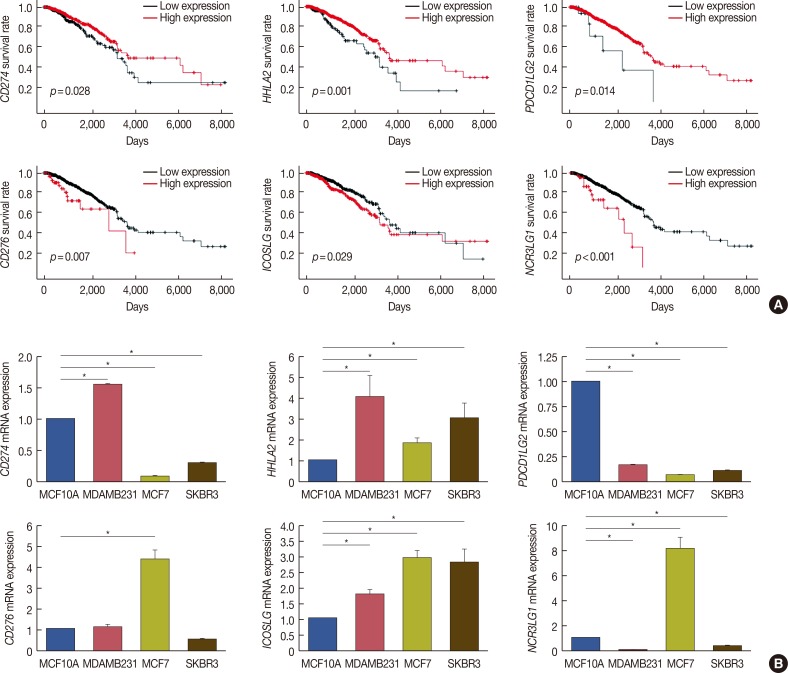
Figure 3
Identify the microRNA may target the B7 family in breast cancer. (A) Heat map of the log2-fold expression changes of 10 microRNAs in the The Cancer Genome Atlas (TCGA) BRCA database (n=1,092). The horizontal row represents different microRNAs, and the vertical column represents different patients. Green squares indicate increases, and red squares indicate decreases. (B) Venn diagrams depicting five sets of data: TargetScan, miRanda, DIANA TOOLS, miRDB and 256 differentially expressed microRNAs in the TCGA database of breast cancer. (C) Venn diagrams were used to select microRNAs that intersect with B7 family members, which have prognostic value in breast cancer. (D) We analyzed the correlation between target microRNAs and the gene expression of B7 family members. The horizontal axis represents microRNA, the vertical axis represents members of the B7 family (n=1,085) (p<0.05). (E) Relationship between microRNA and overall survival in breast cancer.
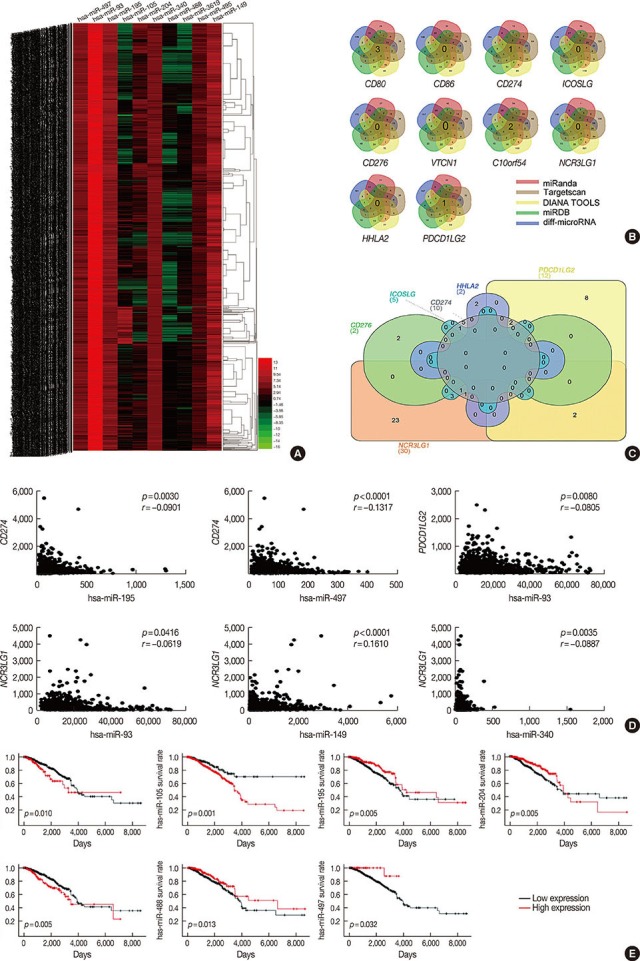
Figure 4
MicroRNA (miR)-195 and miR-497 directly targets CD274. (A) CD274 was detected in MCF7, MDA-MB-231, SK-BR-3 and MCF10A cells by Western blot. (B) Real-time polymerase chain reaction (RT-PCR) was performed to determine miR-195 and miR-497 expression in MCF7, MDA-MB-231, SK-BR-3 and MCF10A. (C) MDA-MB-231 was transfected with miR-195 or miR-497. miR-195, miR-497 and CD274 messenger RNA (mRNA) expression levels were determined via a quantitative RT-PCR assay. (D) MDA-MB-231 was transfected with miR-195 or miR-497. The expression levels of CD274 were determined by Western blotting. (E) CD274 are potential targets of miR-195 and miR-497. The miR-195 and miR-497 target sites and mutant sites in the 3′ untranslated region (3′UTR)s of CD274 are shown. (F) The luciferase vectors that contain the human wild-type (WT) and mutant (MT) CD274 3′UTR regions were co-transfected into MDA-MB-231 cells with miR-195/miR-497 mimic or negative control (NC) miR mimic. The normalized luciferase/Renilla activities were analyzed in the cells 48 hours after the transfection.
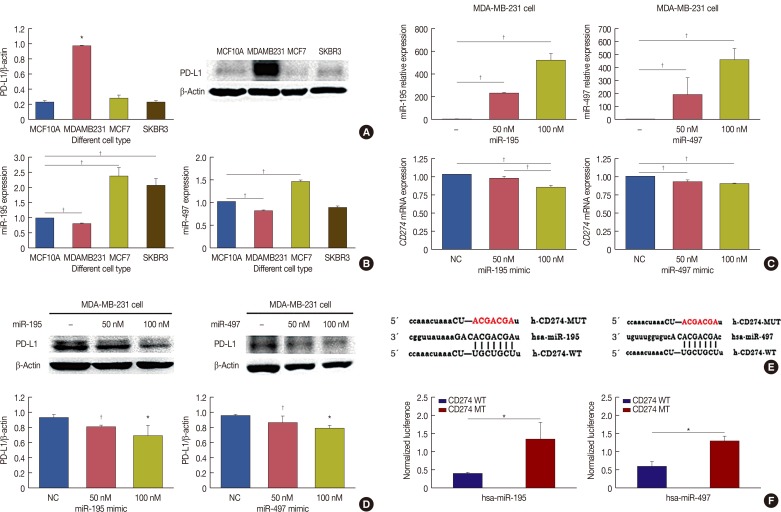
Table 1
Identifying members of the B7 family

CD80=cluster of differentiation 80; CD86=cluster of differentiation 86; CD274=cluster of differentiation 274; PD-L1=programmed death-ligand 1; ICOSLG=inducible T-cell costimulator ligand; CD276=cluster of differentiation 276; VTCN1=V-set domain containing T cell activation inhibitor 1; C10orf54=chromosome 10 open reading frame 54; NCR3LG1=natural killer cell cytotoxicity receptor 3 ligand 1; HHLA2=HERV-H LTR-associating 2; PDCD1LG2=programmed cell death 1 ligand 2.
Table 2
MicroRNAs with more than four intersections

CD80=cluster of differentiation 80; CD86=cluster of differentiation 86; CD274=cluster of differentiation 274; ICOSLG=inducible T-cell costimulator ligand; CD276=cluster of differentiation 276; VTCN1=V-set domain containing T cell activation inhibitor 1; C10orf54=chromosome 10 open reading frame 54; NCR3LG1=natural killer cell cytotoxicity receptor 3 ligand 1; HHLA2=HERV-H LTR-associating 2; PDCD1LG2=programmed cell death 1 ligand 2.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download