Although
Enterococcus faecalis is not commonly considered a microorganism of the normal oral microbiota, it is frequently isolated from obturated root canals of teeth associated with chronic periodontitis, which indicates its pathogenic role in chronic endodontic treatment failure. The gastrointestinal and genitourinary tracts, but not the oral cavity, are recognized as typical habitats of this organism, but the origin of enterococcal infections in root canals is unclear. Some evidence points to exogenous sources; however, the role of normal host microflora cannot be ruled out [
1].
Different virulence factors, including the enterococcal surface protein (Esp), collagen-binding protein (Ace), endocarditis- and biofilm-associated pili (Ebp), cytolysin (Cyl), gelatinase (GelE), bile salt hydrolase, capsule production, and biofilm formation [
2] contribute to the pathogenicity of this organism.
Ace mediates
E. faecalis adherence to human collagen type IV, mouse laminin and human dentin. The
ebp locus, which encodes pili, is well known for its role in endocarditis, but its role in endodontic diseases is unclear.
E. faecalis pathogenicity is also reinforced by biofilm formation, which leads to chronic infections, increased antimicrobial resistance, and protection from the host immune response, and endodontic treatment failure [
2].
As the knowledge about the characteristics of E. faecalis strains isolated from root canals may guide new strategies for treatment of related infections, this study was designed to investigate phenotypic and genotypic characteristics of such isolates. Over a period of 12 months (from May 2016 to May 2017), 70 samples were collected from canals of root-filled teeth (associated with apical periodontitis) of seventy patients seen at the Dentistry School of the Kerman University of Medical Sciences or in private endodontic clinics. The study was approved by the ethics committee of the Bam University of Medical Sciences (Opinion number: MUBAM.REC.1395.3) and was conducted in accordance with the Declaration of Helsinki (2008). Written informed consent was obtained from all patients. All teeth had been root filled (at least one year ago) and apical periodontitis was confirmed based on radiographic evidence. Failure of root-canal treatment was confirmed by an endodontist via clinical and radiographic examinations. Patients who underwent antibiotic therapy within the last three months prior to the study or had systemic diseases were excluded.
The sampling procedure was carried out as follows: After removal of the coronal restorations, resin posts and carious lesions and access cavity preparation, the affected tooth was individually separated from the oral cavity using a rubber dam and disinfected using 5.25% sodium hypochlorite solution inactivated with 5% sodium thiosulfate, in order to avoid interference with the bacterial sampling. A single root canal of each patient was sampled, in order to limit the microbiological assessment to a single ecological environment. In multi-rooted teeth, we sampled the root with the periapical lesion, and if all roots had periapical lesions, the widest canal was selected. The root pulp was removed using Gates-Glidden drills and endodontic files without the use of chemical solvents. Irrigation with sterile saline solution was carried out in order to remove any remaining materials and to moisten the canal prior to sample collection [
3]. The sample was then obtained by introducing a sterile paper point into the full length of the canal (as determined with a preoperative radiograph), and kept in place for 60 s. Next, the introduced paper points were transferred to microtubes containing liquid
Streptococcus faecalis (SF) medium (BBL, Sparks, NV, United States), which were incubated for 48 h at 37ºC. After bacterial growth (characterized by color change of the medium), the bacterial pellet was collected, and identification was initially performed using conventional methods [
4].
E. faecalis identification was confirmed by a polymerase chain reaction (PCR) assay targeting the
ddlE. faecalis gene [
4]. The disk diffusion method was used to test susceptibility to antibiotics. Multidrug-resistance (MDR) was defined as resistance to three or more different classes of antibiotics [
56]. Gelatinase production was assessed using the method described by Marra et al. with minor modifications [
7]. Biofilm formation was assessed by the microtiter plate assay proposed by Upadhyaya et al. [
8], with
Staphylococcus epidermidis (strain ATCC 35984 / RP62A) being used as the positive control. All the assays were performed in triplicate.
Genomic DNA was extracted using an appropriate extraction kit (Gene All, Seoul, Korea), according to the manufacturer's instruction. PCR assays were carried out using specific primer pairs for the e
sp,
gelE,
cylA,
ebp,
ace,
cbh and
cps operons, as previously described (
Table 1) [
491011]. The
cps operon encodes the polysaccharide capsule and has 11 open reading frames (from
cpsA to
cpsK) and 3 polymorphisms: CPS 1, comprising
cpsA and
cpsB; CPS 2, comprising all 11 genes in the operon; and CPS 5, comprising all genes, except
cpsF. Only CPS 2 and 5 encode the capsular polysaccharide [
9].
E. faecalis was isolated from 22 out of 70 specimens and all isolates were susceptible to vancomycin, teicoplanin, and linezolid. MDR was detected in 4/22 (18%) isolates. Antimicrobial susceptibility results are shown in
Table 2. Gelatinase production was detected in 7/22 (32%) isolates. A total of 18 (~80%) isolates were biofilm producers, among which 16 (89%) produced strong biofilm. Optical density (OD) ≥0.24 was considered indicative of strong biofilm formation [
4]. The most prevalent virulence genes were
cbh and
ace each with 72.7%. No
esp-positive isolates were found. The frequency of virulence-associated genes is shown in
Table 2. Amplification of the
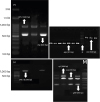
cps operon (
Fig.1) revealed that CPS 1 was the most common polymorphism, expressed by 63% of the isolates, while only two isolates (9%) expressed CPS2.
High prevalence of E. faecalis in treated root canals of teeth associated with periodontitis emphasizes the role of this microorganism in endodontic treatment failures. In this study, 22 E. faecalis isolates from root canals associated with apical periodontitis were analyzed in terms of antimicrobial resistance, biofilm formation, gelatinase production, expression of virulence-associated genes, and capsule locus polymorphism.
Inside root canals,
E. faecalis is protected against the action of antibiotics and the immune system. Beta-lactam antibiotics, such as ampicillin, are used for treating dental infections. It has been reported that ampicillin resistance is not usual in
E. faecalis isolated from patients with periodontitis [
12]. This is corroborated by our results showing an ampicillin-susceptibility rate of 91%.
Up to 40% of
E. faecalis strains isolated from canals of root-filled teeth associated with apical periodontitis were found to be capsule producers in previous reports [
913]. The polysaccharide capsule can hide bacterial surface antigens and render the bacteria resistant to opsonophagocytosis, providing the bacteria a mechanism to evade the inflammatory response in the tissues [
14]. In the present research, most isolates expressed the CPS type 1 polymorphism, thus lacking essential genes of the
cps operon that encode the polysaccharide capsule.
A high proportion of the isolates were shown to produce biofilm, and multiple virulence-associated genes were detected in individual isolates. A study by Sun et al. found that all
E. faecalis strains isolated from patients with chronic periodontitis were able to produce biofilm
in vitro [
15]. Regarding virulence genes, the role of the
esp gene in the
E. faecalis adaptation to the root canal environment has been reported [
13]. In the present study, none of the isolates expressed this gene, which is in contrast with studies reporting an expression rate of 30% and 61% in strains recovered from treated root canals of teeth with or without apical periodontitis [
116]. Cytolysin can cause damage to the dentin and preapical tissues via erythrocyte lysis and destruction of host cells. Virulence determinants such as cytolysin are rare in endodontic isolates and previous reports suggest that cytolysin may have little or no relevance to the pathogenicity of
E. faecalis in endodontic infections [
13]. Herein, 63.6% of the isolates expressed the
cylA gene, but none of them was hemolytic. Our rate was higher than the 38% rate reported by Sun et al. [
15], among which 17% were hemolytic isolates. This lack of phenotypic/genotypic concordance may suggest the loss of the responsible gene in this operon [
10]. Another putative virulence determinant is the
gelE gene, which encodes gelatinase. Gelatinase hydrolyzes gelatin and other peptides, leading to direct and indirect damages to the host tissues and is possibly involved in the biofilm formation on dentin [
1718]. Gelatinase contributes to periodontal inflammation by degradation of specific host proteins and disruption of the periodontal organic matrix [
13]. The present study revealed that 13 (31.8%) dental isolates produced gelatinase, which is in accordance with previous studies [
11516]. Such observations demonstrate that gelatinase can affect the
E. faecalis virulence and, consequently, the pathogenesis of apical and marginal periodontitis [
15]. This lack of genotypic/phenotypic concordance has been reported in other studies [
17]. Expression of the
gelE gene is regulated by a quorum-sensing system encoded by the
fsr gene cluster. A deletion in the upstream sequence of the fsr gene cluster determines the gelatinase-negative phenotype of some
E. faecalis isolates [
19].
The fact that most isolates were biofilm producers suggests biofilm formation may be considered as the most important virulence factor involved in the pathogenesis of enterococcal infections and biofilm eradication should be targeted in therapy strategies.






 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download