1. Randall RL. A promise to our patients with metastatic bone disease. Ann Surg Oncol. 2014; 21:4049–4050.

2. Lipton A, Theriault RL, Hortobagyi GN, et al. Pamidronate prevents skeletal complications and is effective palliative treatment in women with breast carcinoma and osteolytic bone metastases: long term follow-up of two randomized, placebo-controlled trials. Cancer. 2000; 88:1082–1090.

3. Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer. 2002; 2:584–593.

4. Roodman GD. Mechanisms of bone metastasis. N Engl J Med. 2004; 350:1655–1664.

5. Hernandez RK, Adhia A, Wade SW, et al. Prevalence of bone metastases and bone-targeting agent use among solid tumor patients in the United States. Clin Epidemiol. 2015; 7:335–345.
6. Ren G, Esposito M, Kang Y. Bone metastasis and the metastatic niche. J Mol Med (Berl). 2015; 93:1203–1212.

7. Fazilaty H, Mehdipour P. Genetics of breast cancer bone metastasis: a sequential multistep pattern. Clin Exp Metastasis. 2014; 31:595–612.

8. Suva LJ, Washam C, Nicholas RW, et al. Bone metastasis: mechanisms and therapeutic opportunities. Nat Rev Endocrinol. 2011; 7:208–218.

9. Sturge J, Caley MP, Waxman J. Bone metastasis in prostate cancer: emerging therapeutic strategies. Nat Rev Clin Oncol. 2011; 8:357–368.

10. Coleman RE. Bone cancer in 2011: prevention and treatment of bone metastases. Nat Rev Clin Oncol. 2011; 9:76–78.
11. Brook N, Brook E, Dharmarajan A, et al. Breast cancer bone metastases: pathogenesis and therapeutic targets. Int J Biochem Cell Biol. 2018; 96:63–78.

12. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011; 147:275–292.

13. Deo RC. Machine learning in medicine. Circulation. 2015; 132:1920–1930.

14. Deo RC, Nallamothu BK. Learning about machine learning: the promise and pitfalls of big data and the electronic health record. Circ Cardiovasc Qual Outcomes. 2016; 9:618–620.

15. Nemzek JA, Hodges AP, He Y. Bayesian network analysis of multi-compartmentalized immune responses in a murine model of sepsis and direct lung injury. BMC Res Notes. 2015; 8:516.

16. Yoo C, Ramirez L, Liuzzi J. Big data analysis using modern statistical and machine learning methods in medicine. Int Neurourol J. 2014; 18:50–57.

17. Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003; 3:537–549.

18. Ottewell PD. The role of osteoblasts in bone metastasis. J Bone Oncol. 2016; 5:124–127.

19. Haider MT, Holen I, Dear TN, et al. Modifying the osteoblastic niche with zoledronic acid in vivo-potential implications for breast cancer bone metastasis. Bone. 2014; 66:240–250.

20. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013; 41:D991–D995.

21. Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002; 32:Suppl. 496–501.

22. Yu J, Smith VA, Wang PP, et al. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004; 20:3594–3603.

23. Bielza C, Larrañaga P. Bayesian networks in neuroscience: a survey. Front Comput Neurosci. 2014; 8:131.

24. Adabor ES, Acquaah-Mensah GK, Oduro FT. SAGA: a hybrid search algorithm for Bayesian Network structure learning of transcriptional regulatory networks. J Biomed Inform. 2015; 53:27–35.

25. Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013; 29:661–663.

26. Nishida K, Ono K, Kanaya S, et al. KEGGscape: a cytoscape app for pathway data integration. F1000Res. 2014; 3:144.

27. Agostinho NB, Machado KS, Werhli AV. Inference of regulatory networks with a convergence improved MCMC sampler. BMC Bioinformatics. 2015; 16:306.

28. Scutari M, Denis JB. Bayesian networks: with examples in R. Boca Raton, FL: CRC Press;2014.
29. Charniak E. Bayesian networks without tears. Al Mag. 1991; 12:50–63.
30. Kendellen MF, Bradford JW, Lawrence CL, et al. Canonical and non-canonical NF-kappaB signaling promotes breast cancer tumor-initiating cells. Oncogene. 2014; 33:1297–1305.

31. Wang Q, Lu F, Lan R. RNA-sequencing dissects the transcriptome of polyploid cancer cells that are resistant to combined treatments of cisplatin with paclitaxel and docetaxel. Mol Biosyst. 2017; 13:2125–2134.

32. Zhang H, Zhang X, Wu X, et al. Interference of Frizzled 1 (FZD1) reverses multidrug resistance in breast cancer cells through the Wnt/beta-catenin pathway. Cancer Lett. 2012; 323:106–113.

33. Nakao T, Iwata T, Hotchi M, et al. Prediction of response to preoperative chemoradiotherapy and establishment of individualized therapy in advanced rectal cancer. Oncol Rep. 2015; 34:1961–1967.

34. Jiang M, Zhuang H, Xia R, et al. KIF11 is required for proliferation and self-renewal of docetaxel resistant triple negative breast cancer cells. Oncotarget. 2017; 8:92106–92118.

35. Shi Y, Hu W, Yin F, et al. Regulation of drug sensitivity of gastric cancer cells by human calcyclin-binding protein (CacyBP). Gastric Cancer. 2004; 7:160–166.

36. Huth HW, Albarnaz JD, Torres AA, et al. MEK2 controls the activation of MKK3/MKK6-p38 axis involved in the MDA-MB-231 breast cancer cell survival: correlation with cyclin D1 expression. Cell Signal. 2016; 28:1283–1291.

37. Gao S, Ge A, Xu S, et al. PSAT1 is regulated by ATF4 and enhances cell proliferation via the GSK3beta/beta-catenin/cyclin D1 signaling pathway in ER-negative breast cancer. J Exp Clin Cancer Res. 2017; 36:179.

38. Mukherjee S, Das SK. Translocator protein (TSPO) in breast cancer. Curr Mol Med. 2012; 12:443–457.

39. Yonemori K, Seki N, Kurahara H, et al. ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci. 2017; 108:124–135.

40. Rajski M, Vogel B, Baty F, et al. Global gene expression analysis of the interaction between cancer cells and osteoblasts to predict bone metastasis in breast cancer. PLoS One. 2012; 7:e29743.

41. Mourskaia AA, Dong Z, Ng S, et al. Transforming growth factor-beta1 is the predominant isoform required for breast cancer cell outgrowth in bone. Oncogene. 2009; 28:1005–1015.

42. Smid M, Wang Y, Zhang Y, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008; 68:3108–3114.

43. Savci-Heijink CD, Halfwerk H, Koster J, et al. A novel gene expression signature for bone metastasis in breast carcinomas. Breast Cancer Res Treat. 2016; 156:249–259.

44. Yeo SK, French R, Spada F, et al. Opposing roles of Nfkb2 gene products p100 and p52 in the regulation of breast cancer stem cells. Breast Cancer Res Treat. 2017; 162:465–477.

45. Kim B, Kim HH, Lee ZH. Alpha-tocopheryl succinate inhibits osteolytic bone metastasis of breast cancer by suppressing migration of cancer cells and receptor activator of nuclear factor-kappaB ligand expression of osteoblasts. J Bone Metab. 2018; 25:23–33.

46. Fredericks WJ, McGarvey T, Wang H, et al. The bladder tumor suppressor protein TERE1 (UBIAD1) modulates cell cholesterol: implications for tumor progression. DNA Cell Biol. 2011; 30:851–864.

47. Naushad SM, Shree Divyya P, Janaki Ramaiah M, et al. Clinical utility of genetic variants of glutamate carboxypeptidase II in predicting breast cancer and prostate cancer risk. Cancer Genet. 2015; 208:552–558.

48. Zhang XH, Wang Q, Gerald W, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009; 16:67–78.

49. Endo-Munoz L, Cumming A, Sommerville S, et al. Osteosarcoma is characterised by reduced expression of markers of osteoclastogenesis and antigen presentation compared with normal bone. Br J Cancer. 2010; 103:73–81.

50. Sadikovic B, Yoshimoto M, Chilton-MacNeill S, et al. Identification of interactive networks of gene expression associated with osteosarcoma oncogenesis by integrated molecular profiling. Hum Mol Genet. 2009; 18:1962–1975.

51. Zhang XH, Wang Q, Gerald W, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009; 16:67–78.

52. Xu J, Acharya S, Sahin O, et al. 14-3-3zeta turns TGF-beta's function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2. Cancer Cell. 2015; 27:177–192.

53. Mourskaia AA, Amir E, Dong Z, et al. ABCC5 supports osteoclast formation and promotes breast cancer metastasis to bone. Breast Cancer Res. 2012; 14:R149.

54. Kimbung S, Kovacs A, Bendahl PO, et al. Claudin-2 is an independent negative prognostic factor in breast cancer and specifically predicts early liver recurrences. Mol Oncol. 2014; 8:119–128.

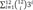 =(3+1)12−1=412−1) different gene configurations g with the collected dataset and using SMILE library (https://www.bayesfusion.com/smile-engine) and C++ program and calculate the following:
=(3+1)12−1=412−1) different gene configurations g with the collected dataset and using SMILE library (https://www.bayesfusion.com/smile-engine) and C++ program and calculate the following: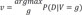 and v′=argmingP(D|V=g)
and v′=argmingP(D|V=g)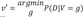 . Note that v and v′ represents 2 different gene expression patterns among the genes in R that maximizes and minimizes P(D|V) respectively. We report the top 4 RR that we calculated from the dataset.
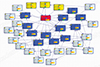
. Note that v and v′ represents 2 different gene expression patterns among the genes in R that maximizes and minimizes P(D|V) respectively. We report the top 4 RR that we calculated from the dataset.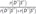 )>99.999% where S′ is the second best CBN). S° includes 1,218 gene expression and a variable called group (representing whether a subject has osteoblast or BM) (Fig. 3). Among 1,219 variables, we selected 59 genes within second degree MB of group (Fig. 4). In addition, CluePedia, one of Cytoscape plugins, identified 13 BCRGs that had common pathways - mammalian target of rapamycin signaling pathway, ErbB signaling pathway, signaling pathways regulating pluripotency of stem cell - in prostate cancers, thyroid cancer and non-small cell lung cancer (Fig. 5).
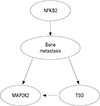
)>99.999% where S′ is the second best CBN). S° includes 1,218 gene expression and a variable called group (representing whether a subject has osteoblast or BM) (Fig. 3). Among 1,219 variables, we selected 59 genes within second degree MB of group (Fig. 4). In addition, CluePedia, one of Cytoscape plugins, identified 13 BCRGs that had common pathways - mammalian target of rapamycin signaling pathway, ErbB signaling pathway, signaling pathways regulating pluripotency of stem cell - in prostate cancers, thyroid cancer and non-small cell lung cancer (Fig. 5). >99.999% where S′ is the second best CBN) (Fig. 6). The first degree MB of group variable was selected from S◆ (we denoted this CBN with 34 variables as S*): 3 genes that were direct causes (parents) of group (NFKB2, UBIAD1, and HEBP1), 12 genes that were direct effects (children) of group (FZD1, MAP2K2, BTNL8, TSPO, HOXB2, FOLH1, KIF11, SLC10A3, PSAT1, CACYBP, S100PBP, and ZFP36L2) and 20 genes that were direct causes (parents) of direct effects (children) of group (BTNL8, TEP1, L1TD1, TSPO, PSMB8, RPS6KB2, DLL3, TNFRSF11B, MAPK3, FASTKD3, ZNF273, PPP2CA, AGA, IFI30, ZDHHC6, INPP1, YWHAB, EPS8, RANBP6, and TATDN2).
>99.999% where S′ is the second best CBN) (Fig. 6). The first degree MB of group variable was selected from S◆ (we denoted this CBN with 34 variables as S*): 3 genes that were direct causes (parents) of group (NFKB2, UBIAD1, and HEBP1), 12 genes that were direct effects (children) of group (FZD1, MAP2K2, BTNL8, TSPO, HOXB2, FOLH1, KIF11, SLC10A3, PSAT1, CACYBP, S100PBP, and ZFP36L2) and 20 genes that were direct causes (parents) of direct effects (children) of group (BTNL8, TEP1, L1TD1, TSPO, PSMB8, RPS6KB2, DLL3, TNFRSF11B, MAPK3, FASTKD3, ZNF273, PPP2CA, AGA, IFI30, ZDHHC6, INPP1, YWHAB, EPS8, RANBP6, and TATDN2).



 PDF
PDF ePub
ePub Citation
Citation Print
Print


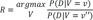









 XML Download
XML Download