1. Yang YX, Metz DC. Safety of proton pump inhibitor exposure. Gastroenterology. 2010; 139:1115–1127.
2. Hagymási K, Müllner K, Herszényi L, Tulassay Z. Update on the pharmacogenomics of proton pump inhibitors. Pharmacogenomics. 2011; 12:873–888.
3. Chaudhry AS, Kochhar R, Kohli KK. Genetic polymorphism of CYP2C19 & therapeutic response to proton pump inhibitors. Indian J Med Res. 2008; 127:521–530.
4. Simon B, Elsner H, Müller P. Protective effect of omeprazole against low-dose acetylsalicylic acid. Endoscopic controlled double-blind study in healthy subjects. Arzneimittelforschung. 1995; 45:701–703.
5. Karam WG, Goldstein JA, Lasker JM, Ghanayem BI. Human CYP2C19 is a major omeprazole 5-hydroxylase, as demonstrated with recombinant cytochrome P450 enzymes. Drug Metab Dispos. 1996; 24:1081–1087.
6. Pearce RE, Rodrigues AD, Goldstein JA, Parkinson A. Identification of the human P450 enzymes involved in lansoprazole metabolism. J Pharmacol Exp Ther. 1996; 277:805–816.
7. VandenBranden M, Ring BJ, Binkley SN, Wrighton SA. Interaction of human liver cytochromes P450 in vitro with LY307640, a gastric proton pump inhibitor. Pharmacogenetics. 1996; 6:81–91.
8. Andersson T. Pharmacokinetics, metabolism and interactions of acid pump inhibitors. Focus on omeprazole, lansoprazole and pantoprazole. Clin Pharmacokinet. 1996; 31:9–28.
9. Meyer UA. Metabolic interactions of the proton-pump inhibitors lansoprazole, omeprazole and pantoprazole with other drugs. Eur J Gastroenterol Hepatol. 1996; 8:Suppl 1. S21–S25.
10. Andersson T, Lagerstrøm PO, Miners JO, Veronese ME, Weidolf L, Birkett DJ. High-performance liquid chromatographic assay for human liver microsomal omeprazole metabolism. J Chromatogr A. 1993; 619:291–297.
11. Li XQ, Weidolf L, Simonsson R, Andersson TB. Enantiomer/enantiomer interactions between the S- and R- isomers of omeprazole in human cytochrome P450 enzymes: major role of CYP2C19 and CYP3A4. J Pharmacol Exp Ther. 2005; 315:777–787.
12. Abelö A, Andersson TB, Antonsson M, Naudot AK, Skånberg I, Weidolf L. Stereoselective metabolism of omeprazole by human cytochrome P450 enzymes. Drug Metab Dispos. 2000; 28:966–972.
13. Ishizaki T, Horai Y. Review article: cytochrome P450 and the metabolism of proton pump inhibitors--emphasis on rabeprazole. Aliment Pharmacol Ther. 1999; 13:Suppl 3. 27–36.
14. Qiao HL, Hu YR, Tian X, Jia LJ, Gao N, Zhang LR, Guo YZ. Pharmacokinetics of three proton pump inhibitors in Chinese subjects in relation to the CYP2C19 genotype. Eur J Clin Pharmacol. 2006; 62:107–112.
15. Hassan-Alin M, Andersson T, Niazi M, Röhss K. A pharmacokinetic study comparing single and repeated oral doses of 20 mg and 40 mg omeprazole and its two optical isomers, S-omeprazole (esomeprazole) and R-omeprazole, in healthy subjects. Eur J Clin Pharmacol. 2005; 60:779–784.
16. Trenk D, Hochholzer W, Fromm MF, Chialda LE, Pahl A, Valina CM, Stratz C, Schmiebusch P, Bestehorn HP, Büttner HJ, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008; 51:1925–1934.
17. Collet JP, Hulot JS, Pena AN, Villard ER, Esteve JB, Cayla G, Silvain G, Beygui F, Payot L, Montalescot G. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: impact on clinical events and on. Eur Heart J. 2009; 30:906.
18. Jeong YH, Kim IS, Kwak CH, Hwang JY. AS-192: the CYP2C19*2 and CYP2C19*3 polymorphisms are associated with high posttreatment platelet reactivity in patients with acute myocardial infarction. Am J Cardiol. 2009; 103:82B.
19. Kim IS, Choi BR, Jeong YH, Kwak CH, Kim S. The CYP2C19*2 and CYP2C19*3 polymorphisms are associated with high post-treatment platelet reactivity in Asian patients with acute coronary syndrome. J Thromb Haemost. 2009; 7:897–899.
20. Marcucci R, Giusti B, Gori A, Paniccia R, Saracini C, Vestrini A, Nanna C, Cordisco A, Antonucci E, Gensini G, et al. Cardiovascular death and nonfatal myocardial infarction in acute coronary syndrome patients are predicted by residual platelet reactivity to ADP in the absence of CYP2C19*2 allele: beyond genetic screening. J Thromb Haemost. 2009; 7:91.
21. Bonello L, Armero S, Ait Mokhtar O, Mancini J, Aldebert P, Saut N, Bonello N, Barragan P, Arques S, Giacomoni MP, et al. Clopidogrel loading dose adjustment according to platelet reactivity monitoring in patients carrying the 2C19*2 loss of function polymorphism. J Am Coll Cardiol. 2010; 56:1630–1636.
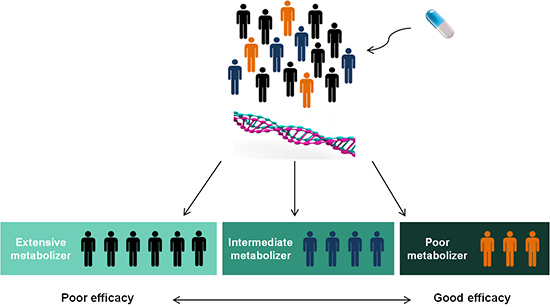
22. Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, Ingelman-Sundberg M. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006; 79:103–113.
23. Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, Rongen GA, van Schaik RH, Schalekamp T, Touw DJ, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011; 89:662–673.
24. Kaneko A, Lum JK, Yaviong L, Takahashi N, Ishizaki T, Bertilsson L, Kobayakawa T, Björkman A. High and variable frequencies of CYP2C19 mutations: medical consequences of poor drug metabolism in Vanuatu and other Pacific islands. Pharmacogenetics. 1999; 9:581–590.
25. Prichard PJ, Yeomans ND, Mihaly GW, Jones DB, Buckle PJ, Smallwood RA, Louis WJ. Omeprazole: a study of its inhibition of gastric pH and oral pharmacokinetics after morning or evening dosage. Gastroenterology. 1985; 88:64–69.
26. Andersson T, Cederberg C, Regårdh CG, Skånberg I. Pharmacokinetics of various single intravenous and oral doses of omeprazole. Eur J Clin Pharmacol. 1990; 39:195–197.
27. Andersson T, Hassan-Alin M, Hasselgren G, Röhss K, Weidolf L. Pharmacokinetic studies with esomeprazole, the (S)-isomer of omeprazole. Clin Pharmacokinet. 2001; 40:411–426.
28. Hofmann U, Schwab M, Treiber G, Klotz U. Sensitive quantification of omeprazole and its metabolites in human plasma by liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2006; 831:85–90.
29. Shimizu M, Uno T, Niioka T, Yaui-Furukori N, Takahata T, Sugawara K, Tateishi T. Sensitive determination of omeprazole and its two main metabolites in human plasma by column-switching high-performance liquid chromatography: application to pharmacokinetic study in relation to CYP2C19 genotypes. J Chromatogr B Analyt Technol Biomed Life Sci. 2006; 832:241–248.
30. Bedada W, de Andrés F, Engidawork E, Pohanka A, Beck O, Bertilsson L, Llerena A, Aklillu E. The psychostimulant Khat (Catha edulis) inhibits CYP2D6 enzyme activity in humans. J Clin Psychopharmacol. 2015; 35:694–699.
31. Sagar M, Seensalu R, Tybring G, Dahl ML, Bertilsson L. CYP2C19 genotype and phenotype determined with omeprazole in patients with acid-related disorders with and without Helicobacter pylori infection. Scand J Gastroenterol. 1998; 33:1034–1038.
32. Furuta T, Ohashi K, Kosuge K, Zhao XJ, Takashima M, Kimura M, Nishimoto M, Hanai H, Kaneko E, Ishizaki T. CYP2C19 genotype status and effect of omeprazole on intragastric pH in humans. Clin Pharmacol Ther. 1999; 65:552–561.
33. Jin SK, Kang TS, Eom SO, Kim JI, Lee HJ, Roh J. CYP2C19 haplotypes in Koreans as a marker of enzyme activity evaluated with omeprazole. J Clin Pharm Ther. 2009; 34:437–446.
34. Cho JY, Yu KS, Jang IJ, Yang BH, Shin SG, Yim DS. Omeprazole hydroxylation is inhibited by a single dose of moclobemide in homozygotic EM genotype for CYP2C19
. Br J Clin Pharmacol. 2002; 53:393–397.
35. Sagar M, Tybring G, Dahl ML, Bertilsson L, Seensalu R. Effects of omeprazole on intragastric pH and plasma gastrin are dependent on the CYP2C19 polymorphism. Gastroenterology. 2000; 119:670–676.
36. Lind T, Cederberg C, Ekenved G, Haglund U, Olbe L. Effect of omeprazole--a gastric proton pump inhibitor--on pentagastrin stimulated acid secretion in man. Gut. 1983; 24:270–276.
37. Andersson T, Regårdh CG. Pharmacokinetics of omeprazole and metabolites following single intravenous and oral doses of 40 and 80mg. Drug Investig. 1990; 2:255–263.
38. Zhou Q, Yamamoto I, Fukuda T, Ohno M, Sumida A, Azuma J. CYP2C19 genotypes and omeprazole metabolism after single and repeated dosing when combined with clarithromycin. Eur J Clin Pharmacol. 1999; 55:43–47.
39. Armstrong D. Review article: gastric pH -- the most relevant predictor of benefit in reflux disease? Aliment Pharmacol Ther. 2004; 20:Suppl 5. 19–26.








 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download