Q fever, a zoonosis caused by
Coxiella burnetii (an obligate intracellular bacterium), occurs worldwide. Not only cattle, sheep, and goats, but also other species (birds, arthropods, and ticks) may also act as reservoirs for
C. burnetii.
C. burnetii can survive for months in the environment; thus, transmission to humans can occur through direct (ingestion or skin inoculation) and indirect (aerosol inhalation) contact of contaminated environs (
1). This caused several large outbreaks of Q fever in Europe (
2). Q fever in humans is characterized by a nonspecific febrile illness that might be accompanied by various degrees of pneumonia or hepatitis. Since Q fever was first described in 1937 by Derrick, it has been considered an under recognized infectious disease because the nonspecific symptoms pose a diagnostic challenge (
3). Previous serologic and bacteriologic studies have suggested that
C. burnetii might be extensively distributed among host animals in Korea (
45). Recently, reports of Q fever in Korea have increased, based on serologic test results (
6). However, the microbiologic characteristics of
C. burnetii have not been reported in Korean patients with Q fever; this is necessary to understand the route of infection and epidemiologic risk. To determine the detailed relationship between
C. burnetii from different geographic origins and hosts, we present the first molecular analysis of C.
burnetii from a patient with Q fever in Korea.
A previously healthy 32-year-old man, an office worker living on the outskirts of Cheongju, Korea was hospitalized for an acute febrile illness in March 2016. He presented with a 5-day history of fever and headache. Physical examination of the chest, abdomen, and skin was initially unremarkable. On admission, his vital signs were: body temperature, 39.6°C; blood pressure, 140/80 mmHg; heart rate, 88 beats/min; and respiratory rate, 20 breaths/min. Complete blood count revealed a normal platelet (217 × 103/µL) and white blood cell (5,720/µL) count, with 77% neutrophils and 17% lymphocytes. Chemistry test results showed elevated levels of C-reactive protein (8.27 mg/dL), aspartate aminotransferase (71 IU/L), and alanine transaminase (76 IU/L). Computed tomography revealed multiple, sub-centimeter lymph nodes in the porta hepatis, inter-aortocaval, and para-aortic areas. Other laboratory and imaging findings were within normal limits. Intravenous ceftriaxone 2 g per day was administered as empiric antibiotic treatment of the febrile illness. There was no bacterial or fungal growth from blood samples obtained for culture prior to antibiotic administration. Because of prolonged fever (> 7 days) despite antibiotic therapy, a serum sample was collected from the patient for specific C. burnetii antibody and nucleic acid detection on hospital day 4. However, he had no history of animal contact. The patient was discharged in an afebrile state after 9 days in hospital as his laboratory findings had normalized.
We used an indirect immunofluorescence antibody (IFA) assay from a commercial kit (IF0200G, IF0200M; Focus Diagnostics, Cyprus, CA, USA) to examine specific antibodies to C. burnetii. The test was performed according to the manufacturer's instructions. First, the serum sample was screened at a 1:16 dilution against C. burnetii phase I and phase II antigens. In sequence, positive samples were diluted twofold from 1:16 to 1:2,048. Diagnosis of acute Q fever is serologically defined as a fourfold rise in the titer of phase II immunoglobulin G (IgG) from serum in the acute phase to the convalescent phase. We confirmed seroconversion between paired serum samples: the initial serum sample (taken on hospital day 4) demonstrated negative results in phase II IgM and IgG; whereas in the serum sample obtained 9 weeks later, the phase II IgG and IgM titers had increased to ≥ 1:2,048 and 1:16, respectively.
Initially, no
C. burnetii specific genes (16S rRNA,
IS1111, and
outer membrane protein [
OMP]) were detected in the whole blood samples. After bacterial amplification of
C. burnetii through animal experiments, nucleic acid of
C. burnetii specific genes
IS1111,
OMP, and 16S rRNA were detected using polymerase chain reaction (PCR); the genes were confirmed as
C. burnetii specific strains using sequencing results. Four-week-old Balb/c mice were purchased from Orientbio (Seoul, Korea). All animals were maintained under Animal Biosafety Level 3 (ABL-3) conditions. All appropriate guidelines for the use and handling of infected animals were followed for the animals infected with
C. burnetii. Buffy coats of
C. burnetii-infected specimens were inoculated into immune-depressed Balb/c mice via the intraperitoneal route. On post-inoculation day 49, the spleens, livers, and lungs of the mice were harvested. Genomic DNA was extracted from inoculated mouse spleens using G-spin Total DNA Extraction Mini kit (Intronbio, Seongnam, Korea), according to the manufacturer's protocol. The extracted DNA specimens were stored at −20°C until analyzed. Nested PCR (nPCR) was used to amplify 16S rRNA of
C. burnetii. First-round PCR was performed with the primers Cox16SF1 (50-CGTAGGAATCTACCTTRTAGWGG-30) and Cox16SR2 (50-GCCTACCCGC TTCTGGTACAATT-30), which produced amplicons with 1,321–1,429 bp. Then, nPCR was performed using the primers Cox16SF2 (50-TGAGAACTAGCTGTTGGRRAGT-30) and Cox16SR2, which produced amplicons with 624–627 bp (
7). Amplicons obtained from nPCR using the
Coxiella specific primers Cox16F2 and Cox16R2 were analyzed using CLC Main Workbench 7.6.4 software (CLC Bio, Aarhus, Denmark) by Jukes-Cantor/Neighbor Joining algorithms. Sequences of
Coxiella 16S rRNA were aligned to determine homology. The stability of the proposed branching order was assessed by bootstrapping (1,000 replicates). The sequence of
C. burnetii 16S rRNA obtained from the human patient's blood and replicated in Balb/c mice was deposited in GenBank (KX825917). The
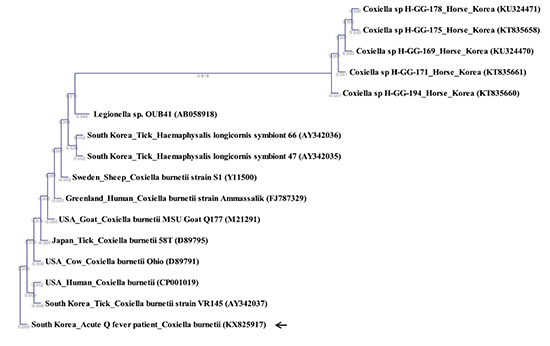
C. burnetii 16S rRNA sequence was compared to published sequences from GenBank. Phylogenetic analysis (Jukes-Cantor/Neighbor Joining) showed high similarity (99.6%–100%) with other strains of
C. burnetii 16S rRNA sequences (
Fig. 1). The sequence results were identical to those obtained from ticks in Korea (AY342037). Compared with
C. burnetii 16S rRNA nucleotides (Y11500, D89791, D89795, FJ787329, M21291, and CP001019) and in view of regional variation, 16S rRNA sequences of this human case in Korea were different from the following
C. burnetii strains: USA, Japan, Greenland, and Sweden (Y11500, D89791, D89795, FJ787329, M21291, and CP001019) (
Fig. 2).
This is the first report to identify the nucleotide sequence of 16S rRNA from a patient with acute Q fever in Korea. Since first described in 1992, there have been reports describing the clinical characteristics of Q fever in Korea (
8910).
C. burnetii has been isolated from the raw milk of dairy cows and has been detected in ticks (
111213). However, the diagnosis of Q fever in humans has mostly been achieved only via serologic testing (such as IFA assays), except in cases of chronic endocarditis, where the diagnosis has been confirmed by nucleic acid detection (
14). Isolation of
C. burnetii is not routinely recommended because it is a difficult and dangerous process. Furthermore, the PCR test performed on clinical specimens has low sensitivity (5%–24%) in acute Q fever and specimens must be obtained within the first 2 weeks of symptom onset and before antibiotic administration (
15). Hence, the microbiologic characteristics of
C. burnetii in Korean patients has previously not been studied. However, it is essential to understand the microbiologic characteristics of
C. burnetii—obtained by genetic analysis—to gain insight into the route of infection or epidemiologic risk. Although epidemiologic investigation showed that dairy goat and dairy sheep farms might lead to large outbreaks of human Q fever in the Netherlands, genetic analysis of
C. burnetii played a key role in elucidating the sources of Q fever outbreaks (
1617).
In Korea, phylogenetic analysis of 16S rRNA sequences for C. burnetii has not previously been reported. In our study, we investigated the origin of Q fever in our patient by comparing the 16S rRNA sequence isolated with those isolated from other countries and hosts. The results of our phylogenetic analysis suggest that the source of Q fever in this case from Korea may not have been from a foreign country.
In our study, phylogenetic analysis of the 16S rRNA nucleotide sequence demonstrated that it was not related to the nucleotide sequences of
Coxiella-like bacteria (CLB) in horses and ticks. A few cases of CLB identified from horses and ticks have been reported in Korea (
813). A previous study showed that the sequences of CLB 16S rRNA were closely related to
C. burnetii, indicating diversity within the genus
Coxiella (
18). Despite genetic similarities, CLB is genetically and etiologically distinct from
C. burnetii. Although a few cases of Q fever in humans in Korea were linked to livestock such as goat and cattle, the primary source of
C. burnetii was uncertain in most cases (
19). Ticks can transmit both
C. burnetii and CLB; hence, they might be a source leading to the emergence of Q fever (
20). Therefore, it is necessary to confirm differences between our human isolate and CLB in Korea.
C. burnetii may be more widespread in South Korea than previously reported. To understand the mode of transmission to humans, further attempts to cultivate C. burnetii are needed from animals, ticks, and environmental samples across Korea, and from more patients. Moreover, we believe that additional investigations into the pathogenicity and infectivity of these isolates are required to complete its characterization.