INTRODUCTION
MATERIALS AND METHODS
Cell cultures and reagents
Cell viability analysis
Wound-healing migration assay
Invasion assay
Western blot analysis
siRNA transfection
Microarray analysis
Gene expression analysis using oligonucleotide microarrays
Cutoffs
Data acquisition and analysis
Patient cohort and tissue microarray (TMA) construction
Immunohistochemistry
Statistical analysis
RESULTS
Western blot of mTOR pathway expression after treatment with triple siRNAs or rapamycin in HG-UC cell lines
 | Fig. 1Protein-level validation of p70S6K and eIF4E suppression after RPS6KB1 and eIF4E siRNA treatments, gene filtering, and gene expression profiling in HG-UC cell lines. (A) Reduced gene expression was observed in 5637 and T24 cells after transfection with siRNAs against p70S6K and eIF4E. Inhibited gene expression of both p70S6K and eIF4E was evident after treatment with triple siRNAs (p70S6K, S6K, and eIF4E). (B) Gene filtering process in 5637 and T24 cells. (C) Hierarchical clustering analysis of 5637 and T24 cells; red spots indicate up-regulation, green spots indicate down-regulation, black spots indicate an absence of modulation, and gray spots indicate the absence of values.
siRNA = small interfering RNA, HG = high-grade, UC = urothelial carcinoma, PI3K = phosphoinositide 3-kinase, mTOR = mammalian target of rapamycin.
|
Gene expression patterns after siRNA or rapamycin treatment
Selection of mTOR pathway downstream genes according to gene expression pattern after siRNA or rapamycin treatment
Table 1
Selection of mTOR pathway downstream genes according to the gene expression patterns after siRNAs or rapamycin treatment in HG-UC cell lines

Patient and tumor characteristics and expression of mTOR pathway downstream genes in relation to clinicopathological variables
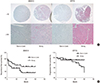 | Fig. 2Expression of downstream genes of the mTOR pathway in HG-UC of bladder tissues, and Kaplan-Meier curves based on staining results in HG-UC patients. (A) Immunohistochemical staining of ANXA10 and ATP7A in paraffin-embedded sections of HG-UC tissues (× 40, × 200). (B) In RFS curves, HG-UC with no or weak ANXA10 immunohistochemical staining exhibited decreased RFS (P = 0.037). In PFS curves, HG-UC with strong ATP7A immunohistochemical staining exhibited decreased PFS (P = 0.004).
mTOR = mammalian target of rapamycin, HG = high-grade, UC = urothelial carcinoma, RFS = recurrence-free survival, PFS = progression-free survival.
|
Table 2
Cox proportional HR to identify predictive factors for recurrence and progression in HG-UC of the bladder

Cell proliferation, wound healing, and invasion inhibition effect of rapamycin in ATP7A knockout 5637 cells
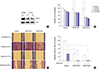 | Fig. 3Down-regulation of up-regulated ATP7A expression after siRNAs or rapamycin treatment in HG-UC 5637 cell line. (A) Western blots of whole-cell lysates from ATP7A stable knockdown and non-targeted shRNA control 5637 cell line after puromycin selection: control, pLKO.1 control vector; shATP7A#1 and shATP7A#2, ATP7A shRNA vectors. (B) Cell viability determined by MTT assay. Data are mean ± SD (n = 6). (C) ATP7A stable knockdown 5637 cell lines exhibited reduced cell migration to rapamycin treatment. (D) ATP7A stable knockdown 5637 cell lines exhibited significantly reduced cell invasion compared to that of the control.
siRNA = small interfering RNA, HG = high-grade, UC = urothelial carcinoma, shRNA = short-hairpin RNA, MTT = 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, SD = standard deviation.
*P < 0.050, †P < 0.010.
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download