1. Bardeesi AS, Gao J, Zhang K, Yu S, Wei M, Liu P, Huang H. A novel role of cellular interactions in vascular calcification. J Transl Med. 2017; 15:95.
2. Proudfoot D, Shanahan CM. Biology of calcification in vascular cells: intima versus media. Herz. 2001; 26:245–251.
3. Farrar DJ, Bond MG, Riley WA, Sawyer JK. Anatomic correlates of aortic pulse wave velocity and carotid artery elasticity during atherosclerosis progression and regression in monkeys. Circulation. 1991; 83:1754–1763.
4. Wallace SM. Yasmin, McEniery CM, Mäki-Petäjä KM, Booth AD, Cockcroft JR, Wilkinson IB. Isolated systolic hypertension is characterized by increased aortic stiffness and endothelial dysfunction. Hypertension. 2007; 50:228–233.
5. Vattikuti R, Towler DA. Osteogenic regulation of vascular calcification: an early perspective. Am J Physiol Endocrinol Metab. 2004; 286:E686–E696.
6. Paloian NJ, Giachelli CM. A current understanding of vascular calcification in CKD. Am J Physiol Renal Physiol. 2014; 307:F891–F900.
7. Albiero M, Avogaro A, Fadini GP. Circulating cellular players in vascular calcification. Curr Pharm Des. 2014; 20:5889–5896.
8. Thompson B, Towler DA. Arterial calcification and bone physiology: role of the bone-vascular axis. Nat Rev Endocrinol. 2012; 8:529–543.
9. Madamanchi NR, Vendrov A, Runge MS. Oxidative stress and vascular disease. Arterioscler Thromb Vasc Biol. 2005; 25:29–38.
10. Schmidt AM, Hori O, Chen JX, Li JF, Crandall J, Zhang J, Cao R, Yan SD, Brett J, Stern D. Advanced glycation endproducts interacting with their endothelial receptor induce expression of vascular cell adhesion molecule-1 (VCAM-1) in cultured human endothelial cells and in mice. A potential mechanism for the accelerated vasculopathy of diabetes. J Clin Invest. 1995; 96:1395–1403.
11. Zhao MM, Xu MJ, Cai Y, Zhao G, Guan Y, Kong W, Tang C, Wang X. Mitochondrial reactive oxygen species promote p65 nuclear translocation mediating high-phosphate-induced vascular calcification in vitro and in vivo. Kidney Int. 2011; 79:1071–1079.
12. Proudfoot D, Skepper JN, Hegyi L, Bennett MR, Shanahan CM, Weissberg PL. Apoptosis regulates human vascular calcification in vitro: evidence for initiation of vascular calcification by apoptotic bodies. Circ Res. 2000; 87:1055–1062.
13. New SE, Aikawa E. Role of extracellular vesicles in de novo mineralization: an additional novel mechanism of cardiovascular calcification. Arterioscler Thromb Vasc Biol. 2013; 33:1753–1758.
14. Yu E, Calvert PA, Mercer JR, Harrison J, Baker L, Figg NL, Kumar S, Wang JC, Hurst LA, Obaid DR, et al. Mitochondrial DNA damage can promote atherosclerosis independently of reactive oxygen species through effects on smooth muscle cells and monocytes and correlates with higher-risk plaques in humans. Circulation. 2013; 128:702–712.
15. Kim H, Kim HJ, Lee K, Kim JM, Kim HS, Kim JR, Ha CM, Choi YK, Lee SJ, Kim JY, et al. a-Lipoic acid attenuates vascular calcification via reversal of mitochondrial function and restoration of Gas6/Axl/Akt survival pathway. J Cell Mol Med. 2012; 16:273–286.
16. Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011; 334:1081–1086.
17. Maltais JS, Simard E, Froehlich U, Denault JB, Gendron L, Grandbois M. iRAGE as a novel carboxymethylated peptide that prevents advanced glycation end product-induced apoptosis and endoplasmic reticulum stress in vascular smooth muscle cells. Pharmacol Res. 2016; 104:176–185.
18. Miyazaki-Anzai S, Masuda M, Demos-Davies KM, Keenan AL, Saunders SJ, Masuda R, Jablonski K, Cavasin MA, Kendrick J, Chonchol M, et al. Endoplasmic reticulum stress effector CCAAT/enhancer-binding protein homologous protein (CHOP) regulates chronic kidney disease-induced vascular calcification. J Am Heart Assoc. 2014; 3:e000949.
19. Serrano RL, Yu W, Terkeltaub R. Mono-allelic and bi-allelic ENPP1 deficiency promote post-injury neointimal hyperplasia associated with increased C/EBP homologous protein expression. Atherosclerosis. 2014; 233:493–502.
20. Duan XH, Chang JR, Zhang J, Zhang BH, Li YL, Teng X, Zhu Y, Du J, Tang CS, Qi YF. Activating transcription factor 4 is involved in endoplasmic reticulum stress-mediated apoptosis contributing to vascular calcification. Apoptosis. 2013; 18:1132–1144.
21. Masuda M, Miyazaki-Anzai S, Levi M, Ting TC, Miyazaki M. PERK-eIF2α-ATF4-CHOP signaling contributes to TNFα-induced vascular calcification. J Am Heart Assoc. 2013; 2:e000238.
22. Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004; 22:233–241.
23. Hruska KA, Mathew S, Saab G. Bone morphogenetic proteins in vascular calcification. Circ Res. 2005; 97:105–114.
24. Nakagawa Y, Ikeda K, Akakabe Y, Koide M, Uraoka M, Yutaka KT, Kurimoto-Nakano R, Takahashi T, Matoba S, Yamada H, et al. Paracrine osteogenic signals via bone morphogenetic protein-2 accelerate the atherosclerotic intimal calcification in vivo. Arterioscler Thromb Vasc Biol. 2010; 30:1908–1915.
25. Derwall M, Malhotra R, Lai CS, Beppu Y, Aikawa E, Seehra JS, Zapol WM, Bloch KD, Yu PB. Inhibition of bone morphogenetic protein signaling reduces vascular calcification and atherosclerosis. Arterioscler Thromb Vasc Biol. 2012; 32:613–622.
26. Cai J, Pardali E, Sánchez-Duffhues G, ten Dijke P. BMP signaling in vascular diseases. FEBS Lett. 2012; 586:1993–2002.
27. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997; 89:755–764.
28. Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997; 89:765–771.
29. Engelse MA, Neele JM, Bronckers AL, Pannekoek H, de Vries CJ. Vascular calcification: expression patterns of the osteoblast-specific gene core binding factor alpha-1 and the protective factor matrix gla protein in human atherogenesis. Cardiovasc Res. 2001; 52:281–289.
30. Steitz SA, Speer MY, Curinga G, Yang HY, Haynes P, Aebersold R, Schinke T, Karsenty G, Giachelli CM. Smooth muscle cell phenotypic transition associated with calcification: upregulation of Cbfa1 and downregulation of smooth muscle lineage markers. Circ Res. 2001; 89:1147–1154.
31. Wilkie AO, Tang Z, Elanko N, Walsh S, Twigg SR, Hurst JA, Wall SA, Chrzanowska KH, Maxson RE Jr. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat Genet. 2000; 24:387–390.
32. Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000; 24:391–395.
33. Shimizu T, Tanaka T, Iso T, Doi H, Sato H, Kawai-Kowase K, Arai M, Kurabayashi M. Notch signaling induces osteogenic differentiation and mineralization of vascular smooth muscle cells: role of Msx2 gene induction via Notch-RBP-Jk signaling. Arterioscler Thromb Vasc Biol. 2009; 29:1104–1111.
34. Shao JS, Aly ZA, Lai CF, Cheng SL, Cai J, Huang E, Behrmann A, Towler DA. Vascular Bmp Msx2 Wnt signaling and oxidative stress in arterial calcification. Ann N Y Acad Sci. 2007; 1117:40–50.
35. Towler DA, Bidder M, Latifi T, Coleman T, Semenkovich CF. Diet-induced diabetes activates an osteogenic gene regulatory program in the aortas of low density lipoprotein receptor-deficient mice. J Biol Chem. 1998; 273:30427–30434.
36. Cheng SL, Behrmann A, Shao JS, Ramachandran B, Krchma K, Bello Arredondo Y, Kovacs A, Mead M, Maxson R, Towler DA. Targeted reduction of vascular Msx1 and Msx2 mitigates arteriosclerotic calcification and aortic stiffness in LDLR-deficient mice fed diabetogenic diets. Diabetes. 2014; 63:4326–4337.
37. Johnson ML, Rajamannan N. Diseases of Wnt signaling. Rev Endocr Metab Disord. 2006; 7:41–49.
38. Shao JS, Cheng SL, Pingsterhaus JM, Charlton-Kachigian N, Loewy AP, Towler DA. Msx2 promotes cardiovascular calcification by activating paracrine Wnt signals. J Clin Invest. 2005; 115:1210–1220.
39. Rong S, Zhao X, Jin X, Zhang Z, Chen L, Zhu Y, Yuan W. Vascular calcification in chronic kidney disease is induced by bone morphogenetic protein-2 via a mechanism involving the Wnt/β-catenin pathway. Cell Physiol Biochem. 2014; 34:2049–2060.
40. Tanaka K, Kaji H, Yamaguchi T, Kanazawa I, Canaff L, Hendy GN, Sugimoto T. Involvement of the osteoinductive factors, Tmem119 and BMP-2, and the ER stress response PERK-eIF2α-ATF4 pathway in the commitment of myoblastic into osteoblastic cells. Calcif Tissue Int. 2014; 94:454–464.
41. Zebboudj AF, Imura M, Boström K. Matrix GLA protein, a regulatory protein for bone morphogenetic protein-2. J Biol Chem. 2002; 277:4388–4394.
42. Yao Y, Bennett BJ, Wang X, Rosenfeld ME, Giachelli C, Lusis AJ, Boström KI. Inhibition of bone morphogenetic proteins protects against atherosclerosis and vascular calcification. Circ Res. 2010; 107:485–494.
43. Reynolds JL, Skepper JN, McNair R, Kasama T, Gupta K, Weissberg PL, Jahnen-Dechent W, Shanahan CM. Multifunctional roles for serum protein fetuin-a in inhibition of human vascular smooth muscle cell calcification. J Am Soc Nephrol. 2005; 16:2920–2930.
44. D’Amelio P, Isaia G, Isaia GC. The osteoprotegerin/RANK/RANKL system: a bone key to vascular disease. J Endocrinol Invest. 2009; 32:6–9.
45. Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Lüthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell. 1997; 89:309–319.
46. Bucay N, Sarosi I, Dunstan CR, Morony S, Tarpley J, Capparelli C, Scully S, Tan HL, Xu W, Lacey DL, et al. osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Genes Dev. 1998; 12:1260–1268.
47. Nakashima T, Takayanagi H. New regulation mechanisms of osteoclast differentiation. Ann N Y Acad Sci. 2011; 1240:E13–E18.
48. Davenport C, Harper E, Forde H, Rochfort KD, Murphy RP, Smith D, Cummins PM. RANKL promotes osteoblastic activity in vascular smooth muscle cells by upregulating endothelial BMP-2 release. Int J Biochem Cell Biol. 2016; 77:171–180.
49. Jenuwein T, Allis CD. Translating the histone code. Science. 2001; 293:1074–1080.
50. Jones P, Altamura S, De Francesco R, Gallinari P, Lahm A, Neddermann P, Rowley M, Serafini S, Steinkühler C. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett. 2008; 18:1814–1819.
51. McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci USA. 2000; 97:14400–14405.
52. Eom GH, Nam YS, Oh JG, Choe N, Min HK, Yoo EK, Kang G, Nguyen VH, Min JJ, Kim JK, et al. Regulation of acetylation of histone deacetylase 2 by p300/CBP-associated factor/histone deacetylase 5 in the development of cardiac hypertrophy. Circ Res. 2014; 114:1133–1143.
53. Nebbioso A, Manzo F, Miceli M, Conte M, Manente L, Baldi A, De Luca A, Rotili D, Valente S, Mai A, et al. Selective class II HDAC inhibitors impair myogenesis by modulating the stability and activity of HDAC-MEF2 complexes. EMBO Rep. 2009; 10:776–782.
54. Eom GH, Kook H. Posttranslational modifications of histone deacetylases: implications for cardiovascular diseases. Pharmacol Ther. 2014; 143:168–180.
55. Vega RB, Harrison BC, Meadows E, Roberts CR, Papst PJ, Olson EN, McKinsey TA. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol Cell Biol. 2004; 24:8374–8385.
56. Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004; 24:8467–8476.
57. Weeks KL, Ranieri A, Karaś A, Bernardo BC, Ashcroft AS, Molenaar C, McMullen JR, Avkiran M. β-adrenergic stimulation induces histone deacetylase 5 (HDAC5) nuclear accumulation in cardiomyocytes by B55α-PP2A-mediated dephosphorylation. J Am Heart Assoc. 2017; 6:e004861.
58. Eom GH, Cho YK, Ko JH, Shin S, Choe N, Kim Y, Joung H, Kim HS, Nam KI, Kee HJ, et al. Casein kinase-2α1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation. 2011; 123:2392–2403.
59. Lkhagva B, Kao YH, Chen YC, Chao TF, Chen SA, Chen YJ. Targeting histone deacetylases: a novel therapeutic strategy for atrial fibrillation. Eur J Pharmacol. 2016; 781:250–257.
60. Williams SM, Golden-Mason L, Ferguson BS, Schuetze KB, Cavasin MA, Demos-Davies K, Yeager ME, Stenmark KR, McKinsey TA, Class I. HDACs regulate angiotensin II-dependent cardiac fibrosis via fibroblasts and circulating fibrocytes. J Mol Cell Cardiol. 2014; 67:112–125.
61. Xie M, Kong Y, Tan W, May H, Battiprolu PK, Pedrozo Z, Wang ZV, Morales C, Luo X, Cho G, et al. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation. 2014; 129:1139–1151.
62. Lee HA, Lee DY, Cho HM, Kim SY, Iwasaki Y, Kim IK. Histone deacetylase inhibition attenuates transcriptional activity of mineralocorticoid receptor through its acetylation and prevents development of hypertension. Circ Res. 2013; 112:1004–1012.
63. Burke MA, Cook SA, Seidman JG, Seidman CE. Clinical and mechanistic insights into the genetics of cardiomyopathy. J Am Coll Cardiol. 2016; 68:2871–2886.
64. Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002; 110:479–488.
65. Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, Epstein JA. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest. 2003; 112:863–871.
66. Kee HJ, Sohn IS, Nam KI, Park JE, Qian YR, Yin Z, Ahn Y, Jeong MH, Bang YJ, Kim N, et al. Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation. 2006; 113:51–59.
67. Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006; 113:2579–2588.
68. Gallo P, Latronico MV, Gallo P, Grimaldi S, Borgia F, Todaro M, Jones P, Gallinari P, De Francesco R, Ciliberto G, et al. Inhibition of class I histone deacetylase with an apicidin derivative prevents cardiac hypertrophy and failure. Cardiovasc Res. 2008; 80:416–424.
69. Kee HJ, Eom GH, Joung H, Shin S, Kim JR, Cho YK, Choe N, Sim BW, Jo D, Jeong MH, et al. Activation of histone deacetylase 2 by inducible heat shock protein 70 in cardiac hypertrophy. Circ Res. 2008; 103:1259–1269.
70. Kee HJ, Kook H. Krüppel-like factor 4 mediates histone deacetylase inhibitor-induced prevention of cardiac hypertrophy. J Mol Cell Cardiol. 2009; 47:770–780.
71. Kim MS, Kwon HJ, Lee YM, Baek JH, Jang JE, Lee SW, Moon EJ, Kim HS, Lee SK, Chung HY, et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat Med. 2001; 7:437–443.
72. Zhang L, Chen B, Zhao Y, Dubielecka PM, Wei L, Qin GJ, Chin YE, Wang Y, Zhao TC. Inhibition of histone deacetylase-induced myocardial repair is mediated by c-kit in infarcted hearts. J Biol Chem. 2012; 287:39338–39348.
73. Zhang L, Qin X, Zhao Y, Fast L, Zhuang S, Liu P, Cheng G, Zhao TC. Inhibition of histone deacetylases preserves myocardial performance and prevents cardiac remodeling through stimulation of endogenous angiomyogenesis. J Pharmacol Exp Ther. 2012; 341:285–293.
74. Kee HJ, Kwon JS, Shin S, Ahn Y, Jeong MH, Kook H. Trichostatin A prevents neointimal hyperplasia via activation of Krüppel like factor 4. Vascul Pharmacol. 2011; 55:127–134.
75. Findeisen HM, Gizard F, Zhao Y, Qing H, Heywood EB, Jones KL, Cohn D, Bruemmer D. Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition. Arterioscler Thromb Vasc Biol. 2011; 31:851–860.
76. Choi JH, Nam KH, Kim J, Baek MW, Park JE, Park HY, Kwon HJ, Kwon OS, Kim DY, Oh GT. Trichostatin A exacerbates atherosclerosis in low density lipoprotein receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2005; 25:2404–2409.
77. Azechi T, Kanehira D, Kobayashi T, Sudo R, Nishimura A, Sato F, Wachi H. Trichostatin A, an HDAC class I/II inhibitor, promotes Pi-induced vascular calcification via up-regulation of the expression of alkaline phosphatase. J Atheroscler Thromb. 2013; 20:538–547.
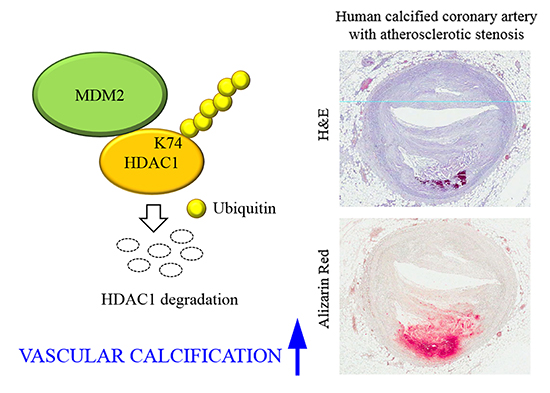
78. Kwon DH, Eom GH, Ko JH, Shin S, Joung H, Choe N, Nam YS, Min HK, Kook T, Yoon S, et al. MDM2 E3 ligase-mediated ubiquitination and degradation of HDAC1 in vascular calcification. Nat Commun. 2016; 7:10492.
79. Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem. 2017; 86:193–224.
80. Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001; 294:1704–1708.
81. Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998; 14:19–57.
82. Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004; 16:119–126.
83. Neutzner M, Neutzner A. Enzymes of ubiquitination and deubiquitination. Essays Biochem. 2012; 52:37–50.
84. Oh YM, Kwon YE, Kim JM, Bae SJ, Lee BK, Yoo SJ, Chung CH, Deshaies RJ, Seol JH. Chfr is linked to tumour metastasis through the downregulation of HDAC1. Nat Cell Biol. 2009; 11:295–302.
85. Gaughan L, Logan IR, Neal DE, Robson CN. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005; 33:13–26.
86. Logan IR, Gaughan L, McCracken SR, Sapountzi V, Leung HY, Robson CN. Human PIRH2 enhances androgen receptor signaling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Mol Cell Biol. 2006; 26:6502–6510.
87. Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E, Ferretti E, Miele E, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010; 12:132–142.
88. Sane S, Rezvani K. Essential roles of E3 ubiquitin ligases in p53 regulation. Int J Mol Sci. 2017; 18:E442.
89. Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992; 358:80–83.
90. Kim BG, Lee JH, Yasuda J, Ryoo HM, Cho JY. Phospho-Smad1 modulation by nedd4 E3 ligase in BMP/TGF-β signaling. J Bone Miner Res. 2011; 26:1411–1424.
91. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016; 17:47–62.
92. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014; 15:509–524.
93. Kim YK. Extracellular microRNAs as biomarkers in human disease. Chonnam Med J. 2015; 51:51–57.
94. Goettsch C, Hutcheson JD, Aikawa E. MicroRNA in cardiovascular calcification: focus on targets and extracellular vesicle delivery mechanisms. Circ Res. 2013; 112:1073–1084.
95. Alkagiet S, Tziomalos K. Vascular calcification: the role of microRNAs. Biomol Concepts. 2017; 8:119–123.
96. Zhou M, Li ZQ, Wang ZL. S100A4 upregulation suppresses tissue ossification and enhances matrix degradation in experimental periodontitis models. Acta Pharmacol Sin. 2015; 36:1388–1394.
97. Choe N, Kwon DH, Shin S, Kim YS, Kim YK, Kim J, Ahn Y, Eom GH, Kook H. The microRNA miR-124 inhibits vascular smooth muscle cell proliferation by targeting S100 calcium-binding protein A4 (S100A4). FEBS Lett. 2017; 591:1041–1052.
98. Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015; 1856:151–164.
99. Carrion K, Dyo J, Patel V, Sasik R, Mohamed SA, Hardiman G, Nigam V. The long non-coding HOTAIR is modulated by cyclic stretch and WNT/β-CATENIN in human aortic valve cells and is a novel repressor of calcification genes. PLoS One. 2014; 9:e96577.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download