1. Chang MS, Lee HS, Kim CW, Kim YI, Kim WH. Clinicopathologic characteristics of Epstein-Barr virus-incorporated gastric cancers in Korea. Pathol Res Pract. 2001; 197:395–400.
2. Fleisher G, Henle W, Henle G, Lennette ET, Biggar RJ. Primary infection with Epstein-Barr virus in infants in the United States: clinical and serologic observations. J Infect Dis. 1979; 139:553–558.
3. Borza CM, Hutt-Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med. 2002; 8:594–599.
4. Okano M, Gross TG. Acute or chronic life-threatening diseases associated with Epstein-Barr virus infection. Am J Med Sci. 2012; 343:483–489.
5. Draborg AH, Duus K, Houen G. Epstein-Barr virus in systemic autoimmune diseases. Clin Dev Immunol. 2013; 2013:535738.
6. Tugizov S, Herrera R, Veluppillai P, Greenspan J, Greenspan D, Palefsky JM. Epstein-Barr virus (EBV)-infected monocytes facilitate dissemination of EBV within the oral mucosal epithelium. J Virol. 2007; 81:5484–5496.
7. Zhu P, Chen YJ, Hao JH, Ge JF, Huang K, Tao RX, Jiang XM, Tao FB. Maternal depressive symptoms related to Epstein-Barr virus reactivation in late pregnancy. Sci Rep. 2013; 3:3096.
8. Elliott SE, Parchim NF, Kellems RE, Xia Y, Soffici AR, Daugherty PS. A pre-eclampsia-associated Epstein-Barr virus antibody cross-reacts with placental GPR50. Clin Immunol. 2016; 168:64–71.
9. Tomai XH. Stillbirth following severe symmetric fetal growth restriction due to reactivation of Epstein-Barr virus infection in pregnancy. J Obstet Gynaecol Res. 2011; 37:1877–1882.
10. Meyohas MC, Maréchal V, Desire N, Bouillie J, Frottier J, Nicolas JC. Study of mother-to-child Epstein-Barr virus transmission by means of nested PCRs. J Virol. 1996; 70:6816–6819.
11. Braz-Silva PH, de Rezende NP, Ortega KL, de Macedo Santos RT, de Magalhães MH. Detection of the Epstein-Barr virus (EBV) by in situ hybridization as definitive diagnosis of hairy leukoplakia. Head Neck Pathol. 2008; 2:19–24.
12. Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006; 7:R100.
13. Iwasaka T, Kidera Y, Tsugitomi H, Sugimori H. The cellular changes in primary and recurrent infection with herpes simplex virus type 2 in an in vitro model. Acta Cytol. 1987; 31:935–940.
14. Juric-Sekhar G, Upton MP, Swanson PE, Westerhoff M. Cytomegalovirus (CMV) in gastrointestinal mucosal biopsies: should a pathologist perform CMV immunohistochemistry if the clinician requests it? Hum Pathol. 2017; 60:11–15.
15. Martínez-López JL, Torres J, Camorlinga-Ponce M, Mantilla A, Leal YA, Fuentes-Pananá EM. Evidence of Epstein-Barr virus association with gastric cancer and non-atrophic gastritis. Viruses. 2014; 6:301–318.
16. Vincent-Bugnas S, Vitale S, Mouline CC, Khaali W, Charbit Y, Mahler P, Prêcheur I, Hofman P, Maryanski JL, Doglio A. EBV infection is common in gingival epithelial cells of the periodontium and worsens during chronic periodontitis. PLoS One. 2013; 8:e80336.
17. Al-Moslih MI, White RJ, Dubes GR. Use of a transfection method to demonstrate a monolayer cell transforming agent from the EB3 line of Burkitt's lymphoma cells. J Gen Virol. 1976; 31:331–345.
18. Guleria I, Khosroshahi A, Ansari MJ, Habicht A, Azuma M, Yagita H, Noelle RJ, Coyle A, Mellor AL, Khoury SJ, et al. A critical role for the programmed death ligand 1 in fetomaternal tolerance. J Exp Med. 2005; 202:231–237.
19. Prasad S, Hu S, Sheng WS, Chauhan P, Singh A, Lokensgard JR. The PD-1: PD-L1 pathway promotes development of brain-resident memory T cells following acute viral encephalitis. J Neuroinflammation. 2017; 14:82.
20. Okazaki T, Honjo T. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 2006; 27:195–201.
21. McNally B, Ye F, Willette M, Flaño E. Local blockade of epithelial PDL-1 in the airways enhances T cell function and viral clearance during influenza virus infection. J Virol. 2013; 87:12916–12924.
22. Delorme-Axford E, Sadovsky Y, Coyne CB. The placenta as a barrier to viral infections. Annu Rev Virol. 2014; 1:133–146.

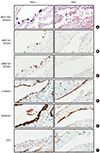

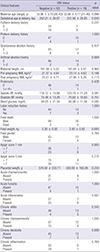
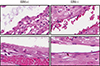





 PDF
PDF ePub
ePub Citation
Citation Print
Print



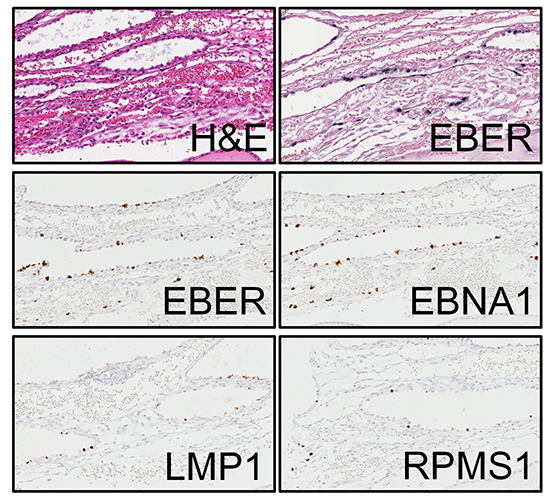
 XML Download
XML Download