Abstract
The initiation of cellular antiviral signaling depends on host pattern-recognition receptors (PRRs)-mediated recognition of viral nucleic acids that are known as classical pathogen-associated molecular patterns (PAMPs). PRRs recruit adaptor proteins and kinases to activate transcription factors and epigenetic modifiers to regulate transcription of hundreds of genes, the products of which collaborate to elicit antiviral responses. In addition, PRRs-triggered signaling induces activation of various inflammasomes which leads to the release of IL-1β and inflammation. Recent studies have demonstrated that PRRs-triggered signaling is critically regulated by ubiquitin and ubiquitin-like molecules. In this review, we first summarize an updated understanding of cellular antiviral signaling and virus-induced activation of inflammasome and then focus on the regulation of key components by ubiquitin and ubiquitin-like molecules.
Go to : 
References
1. Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989; 54:1–13.

3. Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014; 32:461–488.

4. Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005; 6:981–988.

5. Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005; 437:1167–1172.

6. Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005; 19:727–740.
7. Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005; 122:669–682.

8. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013; 339:786–791.

9. Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013; 341:1390–1394.

10. Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009; 138:576–591.

11. Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009; 10:1065–1072.

12. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008; 29:538–550.

13. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008; 455:674–678.

14. Man SM, Karki R, Kanneganti TD. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017; 277:61–75.

15. Yang J, Liu Z, Xiao TS. Post-translational regulation of inflammasomes. Cell Mol Immunol. 2017; 14:65–79.

17. Mevissen TE, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017; 86:159–192.

18. Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015; 16:30–44.

19. dos Santos PF, Mansur DS. Beyond ISGlylation: functions of free intracellular and extracellular ISG15. J Interferon Cytokine Res. 2017; 37:246–253.

21. Xia P, Wang S, Xiong Z, Ye B, Huang LY, Han ZG, Fan Z. IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat Commun. 2015; 6:8132.

22. Yang P, Ma J, Zhang B, Duan H, He Z, Zeng J, Zeng X, Li D, Wang Q, Xiao Y, et al. CpG site-specific hypermethylation of p16INK4α in peripheral blood lymphocytes of PAH-exposed workers. Cancer Epidemiol Biomarkers Prev. 2012; 21:182–190.

24. Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, Wang P, Zhao K, Hou J, Wang X, et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol. 2017; 18:214–224.

25. Wang Q, Liu X, Cui Y, Tang Y, Chen W, Li S, Yu H, Pan Y, Wang C. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014; 41:919–933.

26. Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012; 287:28646–28655.

27. Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010; 33:765–776.

28. Michallet MC, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, Curran J, Poeck H, Bscheider M, Hartmann G, König M, et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008; 28:651–661.

29. Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. ELife. 2013; 2:e00785.

30. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015; 347:aaa2630.

31. Shinohara ML, Lu L, Bu J, Werneck MB, Kobayashi KS, Glimcher LH, Cantor H. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol. 2006; 7:498–506.

32. Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005; 434:772–777.

33. Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005; 434:1035–1040.

34. Lu B, Ren Y, Sun X, Han C, Wang H, Chen Y, Peng Q, Cheng Y, Cheng X, Zhu Q, et al. Induction of INKIT by viral infection negatively regulates antiviral responses through inhibiting phosphorylation of p65 and IRF3. Cell Host Microbe. 2017; 22:86–98. e4.

35. Jin J, Hu H, Li HS, Yu J, Xiao Y, Brittain GC, Zou Q, Cheng X, Mallette FA, Watowich SS, et al. Noncanonical NF-κ B pathway controls the production of type I interferons in antiviral innate immunity. Immunity. 2014; 40:342–354.

36. Mathur A, Hayward JA, Man SM. Molecular mechanisms of inflammasome signaling. J Leukoc Biol.DOI: doi: 10.1189/jlb.3MR0617–250R.
38. Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010; 11:395–402.

39. Singh VV, Kerur N, Bottero V, Dutta S, Chakraborty S, Ansari MA, Paudel N, Chikoti L, Chandran B. Kaposi's sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes. J Virol. 2013; 87:4417–4431.

40. Johnson KE, Chikoti L, Chandran B. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol. 2013; 87:5005–5018.

41. Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschläger N, Schlee M, Rothenfusser S, Barchet W, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010; 11:63–69.

42. Chattergoon MA, Latanich R, Quinn J, Winter ME, Buckheit RW 3rd, Blankson JN, Pardoll D, Cox AL. HIV and HCV activate the inflammasome in monocytes and macrophages via endosomal Toll-like receptors without induction of type 1 interferon. PLoS Pathog. 2014; 10:e1004082.

43. Ogura Y, Sutterwala FS, Flavell RA. The inflammasome: first line of the immune response to cell stress. Cell. 2006; 126:659–662.

44. Oda T, Akaike T, Hamamoto T, Suzuki F, Hirano T, Maeda H. Oxygen radicals in influenza-induced pathogenesis and treatment with pyran polymer-conjugated SOD. Science. 1989; 244:974–976.

45. Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010; 11:404–410.

46. Rahman MM, McFadden G. Myxoma virus lacking the pyrin-like protein M013 is sensed in human myeloid cells by both NLRP3 and multiple Toll-like receptors, which independently activate the inflammasome and NF-κ B innate response pathways. J Virol. 2011; 85:12505–12517.

47. Martin BN, Wang C, Willette-Brown J, Herjan T, Gulen MF, Zhou H, Bulek K, Franchi L, Sato T, Alnemri ES, et al. IKKα negatively regulates ASC-dependent inflammasome activation. Nat Commun. 2014; 5:4977.

48. Wang Y, Ning X, Gao P, Wu S, Sha M, Lv M, Zhou X, Gao J, Fang R, Meng G, et al. Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity. 2017; 46:393–404.

49. Lin D, Zhong B. Regulation of cellular innate antiviral signaling by ubiquitin modification. Acta Biochim Biophys Sin (Shanghai). 2015; 47:149–155.

50. Liu W, Li J, Zheng W, Shang Y, Zhao Z, Wang S, Bi Y, Zhang S, Xu C, Duan Z, et al. Cyclophilin A-regulated ubiquitination is critical for RIG-I-mediated antiviral immune responses. ELife. 2017; 6:e24425.

51. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007; 446:916–920.

52. Shi Y, Yuan B, Zhu W, Zhang R, Li L, Hao X, Chen S, Hou F. Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat Commun. 2017; 8:15138.

53. Sun X, Xian H, Tian S, Sun T, Qin Y, Zhang S, Cui J. A hierarchical mechanism of RIG-I ubiquitination provides sensitivity, robustness and synergy in antiviral immune responses. Sci Rep. 2016; 6:29263.

54. Xian H, Xie W, Yang S, Liu Q, Xia X, Jin S, Sun T, Cui J. Stratified ubiquitination of RIG-I creates robust immune response and induces selective gene expression. Sci Adv. 2017; 3:e1701764.

55. Hao Q, Jiao S, Shi Z, Li C, Meng X, Zhang Z, Wang Y, Song X, Wang W, Zhang R, et al. A non-canonical role of the p97 complex in RIG-I antiviral signaling. EMBO J. 2015; 34:2903–2920.

56. Wang W, Jiang M, Liu S, Zhang S, Liu W, Ma Y, Zhang L, Zhang J, Cao X. RNF122 suppresses antiviral type I interferon production by targeting RIG-I CARDs to mediate RIG-I degradation. Proc Natl Acad Sci U S A. 2016; 113:9581–9586.

57. Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A. 2007; 104:7500–7505.

58. Chen W, Han C, Xie B, Hu X, Yu Q, Shi L, Wang Q, Li D, Wang J, Zheng P, et al. Induction of Siglec-G by RNA viruses inhibits the innate immune response by promoting RIG-I degradation. Cell. 2013; 152:467–478.

59. Zhao C, Jia M, Song H, Yu Z, Wang W, Li Q, Zhang L, Zhao W, Cao X. The E3 ubiquitin ligase TRIM40 attenuates antiviral immune responses by targeting MDA5 and RIG-I. Cell Rep. 2017; 21:1613–1623.

60. Zhang H, Wang D, Zhong H, Luo R, Shang M, Liu D, Chen H, Fang L, Xiao S. Ubiquitin-specific protease 15 negatively regulates virus-induced type I interferon signaling via catalytically-dependent and -independent mechanisms. Sci Rep. 2015; 5:11220.

61. Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, Gack MU. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal. 2014; 7:ra3.

62. Zhang M, Wu X, Lee AJ, Jin W, Chang M, Wright A, Imaizumi T, Sun SC. Regulation of IkappaB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008; 283:18621–18626.

63. Chen R, Zhang L, Zhong B, Tan B, Liu Y, Shu HB. The ubiquitin-specific protease 17 is involved in virus-triggered type I IFN signaling. Cell Res. 2010; 20:802–811.

64. Wang L, Zhao W, Zhang M, Wang P, Zhao K, Zhao X, Yang S, Gao C. USP4 positively regulates RIG-I-mediated antiviral response through deubiquitination and stabilization of RIG-I. J Virol. 2013; 87:4507–4515.

65. Cui J, Song Y, Li Y, Zhu Q, Tan P, Qin Y, Wang HY, Wang RF. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 2014; 24:400–416.

66. Fan Y, Mao R, Yu Y, Liu S, Shi Z, Cheng J, Zhang H, An L, Zhao Y, Xu X, et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med. 2014; 211:313–328.

67. Lang X, Tang T, Jin T, Ding C, Zhou R, Jiang W. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J Exp Med. 2017; 214:459–473.

68. Wang Q, Huang L, Hong Z, Lv Z, Mao Z, Tang Y, Kong X, Li S, Cui Y, Liu H, et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 2017; 13:e1006264.

69. Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, Zhou Y, Chen Y, Huang J, Wang RF, et al. TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol Cell. 2016; 64:105–119.

70. Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012; 13:255–263.

71. Han S, Lear TB, Jerome JA, Rajbhandari S, Snavely CA, Gulick DL, Gibson KF, Zou C, Chen BB, Mallampalli RK. Lipopolysaccharide primes the NALP3 inflammasome by inhibiting its ubiquitination and degradation mediated by the SCFFBXL2 E3 ligase. J Biol Chem. 2015; 290:18124–18133.

72. Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, Zhou R. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015; 160:62–73.

73. Xue Q, Zhou Z, Lei X, Liu X, He B, Wang J, Hung T. TRIM38 negatively regulates TLR3-mediated IFN-β signaling by targeting TRIF for degradation. PLoS One. 2012; 7:e46825.

74. Hu MM, Xie XQ, Yang Q, Liao CY, Ye W, Lin H, Shu HB. TRIM38 negatively regulates TLR3/4-mediated innate immune and inflammatory responses by two sequential and distinct mechanisms. J Immunol. 2015; 195:4415–4425.

75. Yang Y, Liao B, Wang S, Yan B, Jin Y, Shu HB, Wang YY. E3 ligase WWP2 negatively regulates TLR3-mediated innate immune response by targeting TRIF for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2013; 110:5115–5120.

76. Yang Q, Liu TT, Lin H, Zhang M, Wei J, Luo WW, Hu YH, Zhong B, Hu MM, Shu HB. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017; 13:e1006600.

77. Lee BC, Miyata M, Lim JH, Li JD. Deubiquitinase CYLD acts as a negative regulator for bacterium NTHi-induced inflammation by suppressing K63-linked ubiquitination of MyD88. Proc Natl Acad Sci U S A. 2016; 113:E165–E171.

78. Naiki Y, Michelsen KS, Zhang W, Chen S, Doherty TM, Arditi M. Transforming growth factor-beta differentially inhibits MyD88-dependent, but not TRAM- and TRIF-dependent, lipopolysaccharide-induced TLR4 signaling. J Biol Chem. 2005; 280:5491–5495.

79. Strickson S, Campbell DG, Emmerich CH, Knebel A, Plater L, Ritorto MS, Shpiro N, Cohen P. The anti-inflammatory drug BAY 11–7082 suppresses the MyD88-dependent signalling network by targeting the ubiquitin system. Biochem J. 2013; 451:427–437.

80. Ji S, Sun M, Zheng X, Li L, Sun L, Chen D, Sun Q. Cell-surface localization of Pellino antagonizes Toll-mediated innate immune signalling by controlling MyD88 turnover in Drosophila. Nat Commun. 2014; 5:3458.

81. Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. The E3 ubiquitin ligase Nrdp1 ‘preferentially'promotes TLR-mediated production of type I interferon. Nat Immunol. 2009; 10:744–752.

82. Jin S, Tian S, Luo M, Xie W, Liu T, Duan T, Wu Y, Cui J. Tetherin suppresses type I interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol Cell. 2017; 68:308–322. e4.

83. Xing J, Zhang A, Zhang H, Wang J, Li XC, Zeng MS, Zhang Z. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat Commun. 2017; 8:945.

84. Ni G, Konno H, Barber GN. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci Immunol. 2017; 2:eaah7119.

85. Wang Y, Lian Q, Yang B, Yan S, Zhou H, He L, Lin G, Lian Z, Jiang Z, Sun B. TRIM30α is a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLoS Pathog. 2015; 11:e1005012.

86. Zhang M, Zhang MX, Zhang Q, Zhu GF, Yuan L, Zhang DE, Zhu Q, Yao J, Shu HB, Zhong B. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Res. 2016; 26:1302–1319.

87. Sun H, Zhang Q, Jing YY, Zhang M, Wang HY, Cai Z, Liuyu T, Zhang ZD, Xiong TC, Wu Y, et al. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat Commun. 2017; 8:15534.

88. Chen Y, Wang L, Jin J, Luan Y, Chen C, Li Y, Chu H, Wang X, Liao G, Yu Y, et al. p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation. J Exp Med. 2017; 214:991–1010.

89. Mao AP, Li S, Zhong B, Li Y, Yan J, Li Q, Teng C, Shu HB. Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-beta (IFN-beta) and cellular antiviral response. J Biol Chem. 2010; 285:9470–9476.

90. Wang C, Huang Y, Sheng J, Huang H, Zhou J. Estrogen receptor alpha inhibits RLR-mediated immune response via ubiquitinating TRAF3. Cell Signal. 2015; 27:1977–1983.

91. Peng Y, Xu R, Zheng X. HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiquitination via recruiting OTUB1. PLoS Pathog. 2014; 10:e1004041.

92. Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, Gao Y, Ran Y, Tien P, Shu HB. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010; 285:4291–4297.

93. Lin D, Zhang M, Zhang MX, Ren Y, Jin J, Zhao Q, Pan Z, Wu M, Shu HB, Dong C, et al. Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc Natl Acad Sci U S A. 2015; 112:11324–11329.

94. Panda S, Nilsson JA, Gekara NO. Deubiquitinase MYSM1 regulates innate immunity through inactivation of TRAF3 and TRAF6 complexes. Immunity. 2015; 43:647–659.

95. Guan K, Wei C, Zheng Z, Song T, Wu F, Zhang Y, Cao Y, Ma S, Chen W, Xu Q, et al. MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J Immunol. 2015; 194:4880–4890.

96. An T, Li S, Pan W, Tien P, Zhong B, Shu HB, Wu S. DYRK2 negatively regulates type I interferon induction by promoting TBK1 degradation via Ser527 phosphorylation. PLoS Pathog. 2015; 11:e1005179.

97. Wildling L, Unterauer B, Zhu R, Rupprecht A, Haselgrübler T, Rankl C, Ebner A, Vater D, Pollheimer P, Pohl EE, et al. Linking of sensor molecules with amino groups to amino-functionalized AFM tips. Bioconjug Chem. 2011; 22:1239–1248.

98. Song G, Liu B, Li Z, Wu H, Wang P, Zhao K, Jiang G, Zhang L, Gao C. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat Immunol. 2016; 17:1342–1351.

99. Zhao X, Zhu H, Yu J, Li H, Ge J, Chen W. c-Cbl-mediated ubiquitination of IRF3 negatively regulates IFN-β production and cellular antiviral response. Cell Signal. 2016; 28:1683–1693.

100. Chattopadhyay S, Kuzmanovic T, Zhang Y, Wetzel JL, Sen GC. Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity. 2016; 44:1151–1161.

101. Ning S, Campos AD, Darnay BG, Bentz GL, Pagano JS. TRAF6 and the three C-terminal lysine sites on IRF7 are required for its ubiquitination-mediated activation by the tumor necrosis factor receptor family member latent membrane protein 1. Mol Cell Biol. 2008; 28:6536–6546.

102. Decque A, Joffre O, Magalhaes JG, Cossec JC, Blecher-Gonen R, Lapaquette P, Silvin A, Manel N, Joubert PE, Seeler JS, et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol. 2016; 17:140–149.

103. Mi Z, Fu J, Xiong Y, Tang H. SUMOylation of RIG-I positively regulates the type I interferon signaling. Protein Cell. 2010; 1:275–283.

104. Fu J, Xiong Y, Xu Y, Cheng G, Tang H. MDA5 is SUMOylated by PIAS2β in the upregulation of type I interferon signaling. Mol Immunol. 2011; 48:415–422.

105. Doiron K, Goyon V, Coyaud E, Rajapakse S, Raught B, McBride HM. The dynamic interacting landscape of MAPL reveals essential functions for SUMOylation in innate immunity. Sci Rep. 2017; 7:107.

106. Hu MM, Liao CY, Yang Q, Xie XQ, Shu HB. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J Exp Med. 2017; 214:973–989.

107. Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT, Yin L, Shu HB. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 2016; 45:555–569.

108. Cui Y, Yu H, Zheng X, Peng R, Wang Q, Zhou Y, Wang R, Wang J, Qu B, Shen N, et al. SENP7 potentiates cGAS activation by relieving SUMO-mediated inhibition of cytosolic DNA sensing. PLoS Pathog. 2017; 13:e1006156.

109. Sankineni S, Cho Y, Hosseinian N, Kolliputi N. Does pIgR down-regulation in COPD cause reprogramming of bronchial epithelium? Lung. 2015; 193:1–2.
110. Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, Kato A, Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J Biol Chem. 2008; 283:25660–25670.

111. Liang Q, Deng H, Li X, Wu X, Tang Q, Chang TH, Peng H, Rauscher FJ 3rd, Ozato K, Zhu F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J Immunol. 2011; 187:4754–4763.

112. Li R, Pan Y, Shi DD, Zhang Y, Zhang J. PIAS1 negatively modulates virus triggered type I IFN signaling by blocking the DNA binding activity of IRF3. Antiviral Res. 2013; 100:546–554.

113. Lumpkin RJ, Gu H, Zhu Y, Leonard M, Ahmad AS, Clauser KR, Meyer JG, Bennett EJ, Komives EA. Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat Commun. 2017; 8:1171.

114. Lamoliatte F, McManus FP, Maarifi G, Chelbi-Alix MK, Thibault P. Uncovering the SUMOylation and ubiquitylation crosstalk in human cells using sequential peptide immunopurification. Nat Commun. 2017; 8:14109.

115. Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, García-Sastre A, et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A. 2007; 104:1371–1376.

116. Ritchie KJ, Hahn CS, Kim KI, Yan M, Rosario D, Li L, de la Torre JC, Zhang DE. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med. 2004; 10:1374–1378.

117. Ketscher L, Hannß R, Morales DJ, Basters A, Guerra S, Goldmann T, Hausmann A, Prinz M, Naumann R, Pekosz A, et al. Selective inactivation of USP18 isopeptidase activity in vivo enhances ISG15 conjugation and viral resistance. Proc Natl Acad Sci U S A. 2015; 112:1577–1582.

118. Arimoto K, Konishi H, Shimotohno K. UbcH8 regulates ubiquitin and ISG15 conjugation to RIG-I. Mol Immunol. 2008; 45:1078–1084.

119. Lu G, Reinert JT, Pitha-Rowe I, Okumura A, Kellum M, Knobeloch KP, Hassel B, Pitha PM. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell Mol Biol (Noisy-le-grand). 2006; 52:29–41.
120. Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, Zhu LH, Wang C. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol. 2010; 30:2424–2436.

121. Werneke SW, Schilte C, Rohatgi A, Monte KJ, Michault A, Arenzana-Seisdedos F, Vanlandingham DL, Higgs S, Fontanet A, Albert ML, et al. ISG15 is critical in the control of Chikungunya virus infection independent of UbE1L mediated conjugation. PLoS Pathog. 2011; 7:e1002322.

122. Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, Salem S, Radovanovic I, Grant AV, Adimi P, et al. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 2012; 337:1684–1688.

123. Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, et al. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 2015; 517:89–93.

124. Meuwissen ME, Schot R, Buta S, Oudesluijs G, Tinschert S, Speer SD, Li Z, van Unen L, Heijsman D, Goldmann T, et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J Exp Med. 2016; 213:1163–1174.

125. Strebovsky J, Walker P, Lang R, Dalpke AH. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. FASEB J. 2011; 25:863–874.

126. Chang PJ, Chen LW, Chen LY, Hung CH, Shih YJ, Wang SS. Effects of the NEDD8-activating enzyme inhibitor MLN4924 on lytic reactivation of Kaposi's sarcoma-associated herpesvirus. J Virol. 2017; 91:e00505–17.

127. Davis KA, Morelli M, Patton JT. Rotavirus NSP1 requires casein kinase II-mediated phosphorylation for hijacking of cullin-RING ligases. MBio. 2017; 8:e01213–17.

128. Ding S, Mooney N, Li B, Kelly MR, Feng N, Loktev AV, Sen A, Patton JT, Jackson PK, Greenberg HB. Comparative proteomics reveals strain-specific β-TrCP degradation via rotavirus NSP1 hijacking a host cullin-3-Rbx1 complex. PLoS Pathog. 2016; 12:e1005929.

129. Coleman KE, Békés M, Chapman JR, Crist SB, Jones MJ, Ueberheide BM, Huang TT. SENP8 limits aberrant neddylation of NEDD8 pathway components to promote cullin-RING ubiquitin ligase function. ELife. 2017; 6:e24325.

130. Noguchi K, Okumura F, Takahashi N, Kataoka A, Kamiyama T, Todo S, Hatakeyama S. TRIM40 promotes neddylation of IKKγ and is downregulated in gastrointestinal cancers. Carcinogenesis. 2011; 32:995–1004.

131. Yan F, Guan J, Peng Y, Zheng X. MyD88 NEDDylation negatively regulates MyD88-dependent NF-κB signaling through antagonizing its ubiquitination. Biochem Biophys Res Commun. 2017; 482:632–637.

132. Chan Y, Yoon J, Wu JT, Kim HJ, Pan KT, Yim J, Chien CT. DEN1 deneddylates non-cullin proteins in vivo. J Cell Sci. 2008; 121:3218–3223.
133. Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B, Hay RT. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003; 278:25637–25643.

134. Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, Chen A, Lee CG, Wei N, Wilkinson KD, Wang R, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003; 278:28882–28891.

135. Kumari P, Kumar H. Viral deubiquitinases: role in evasion of anti-viral innate immunity. Crit Rev Microbiol.DOI: doi: 10.1080/1040841X.2017.1368999.
136. Zhao K, Zhang Q, Li X, Zhao D, Liu Y, Shen Q, Yang M, Wang C, Li N, Cao X. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J Immunol. 2016; 196:1209–1217.

137. Ye JS, Kim N, Lee KJ, Nam YR, Lee U, Joo CH. Lysine 63-linked TANK-binding kinase 1 ubiquitination by mindbomb E3 ubiquitin protein ligase 2 is mediated by the mitochondrial antiviral signaling protein. J Virol. 2014; 88:12765–12776.

138. Yu Z, Song H, Jia M, Zhang J, Wang W, Li Q, Zhang L, Zhao W. USP1-UAF1 deubiquitinase complex stabilizes TBK1 and enhances antiviral responses. J Exp Med. 2017; 214:3553–3563.

139. Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, Wang RF. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012; 13:387–395.

Go to : 
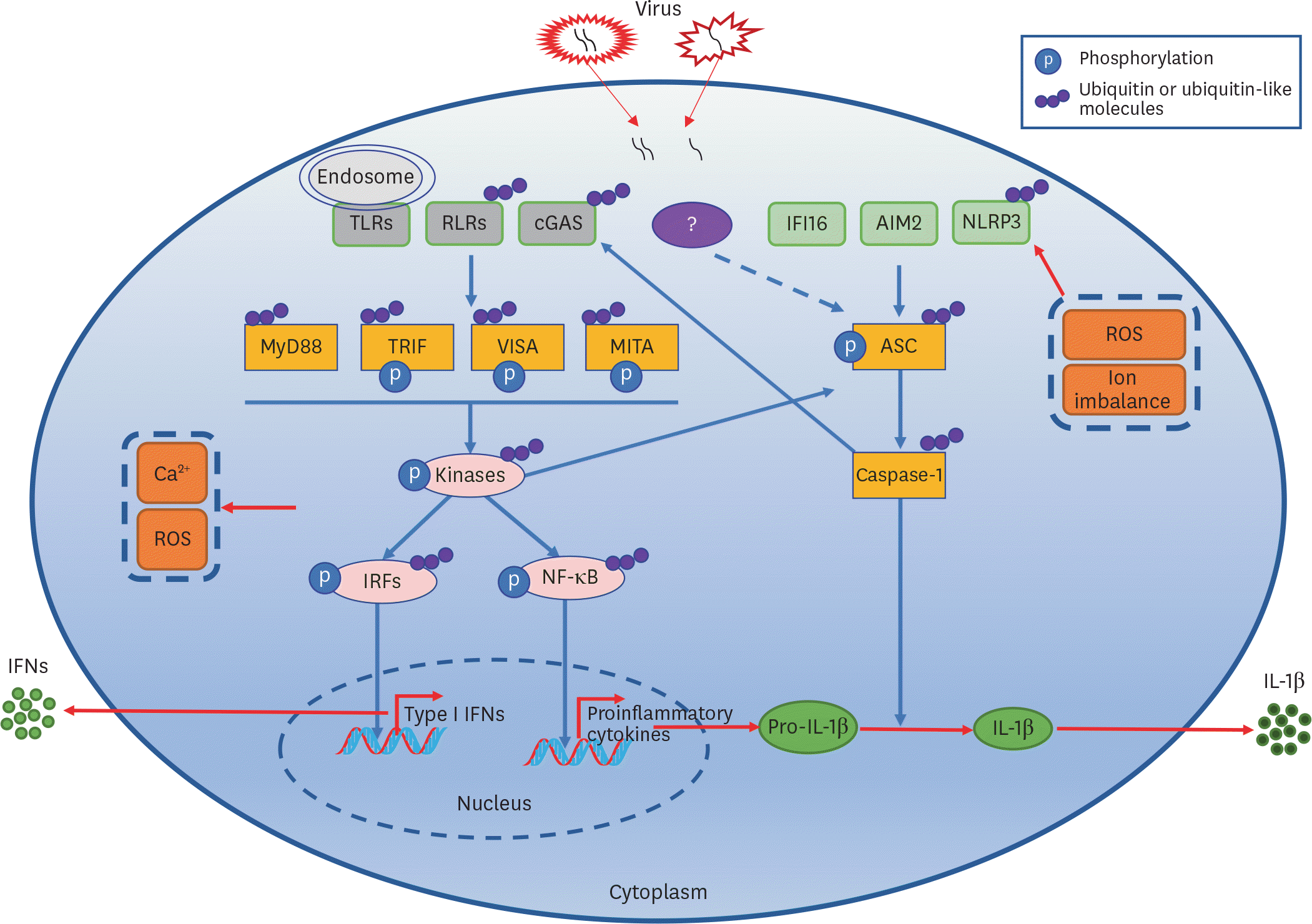 | Figure 1.Virus-triggered PRRs-mediated signaling pathways leading to type I IFN and IL-1β induction. |
Table 1.
Ubiquitin and ubiquitin-like modifications in virus-triggered PRRs-mediated signaling pathways




 PDF
PDF Citation
Citation Print
Print


 XML Download
XML Download