INTRODUCTION
MATERIALS AND METHODS
Randomization of FR codons
Phage enzyme immunoassay
Sequence analysis
Generation of scFv-displaying phages with mutated H82, H82A, and H82B residues
Enzyme immunoassay of chicken-human chimeric IgG1 with mutated H78 and L85 residues
RESULTS
Versatility of the framework residues in chicken scFv
 | Figure 1Comparing the interchangeability of 161 FR residues in PC24 scFv with the natural mutations found in naturally-occurring chicken antibodies. Amino acids were classified into 5 group of amino acid based on their physicochemical properties: [GAPVLIM], blue; [FYW], purple; [STCNQ], green; [KRH], red; [DE], orange. Positive rate was defined as the percentage of positive clones among 92 clones tested in phage enzyme immunoassay. The secondary structure of chicken antibody FR was adopted from the anti-phospho-tau antibody (PDB ID: 4GLR) (18). The sequences of 150 antigen-reactive chicken antibodies were collected, analyzed in the same manner, and presented as the natural mutation. |
Effect of mutating 3 consecutive framework residues
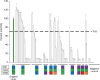 | Figure 2Effects of mutating 3 consecutive residues (H82, H82A, and H82B) on the reactivity of the scFv. With H82A FR residue were replaced into different amino acid in the same group of amino acid ([STCHQ], green), 70 different mutants were created by replacing H82 (A, V, L, F, and W) and H82B (G, P, V, L, F, W, S, T, C, N, Q, K, R, and D) FR residues simultaneously. Phage enzyme immunoassay was performed on the mutant scFv-displaying phage to determine its reactivity. |
Versatility of the framework residues in 2 other chicken scFvs
 | Figure 3Versatility of 16 FR residues in 2 other chicken antibodies (PC17 and VCM2). Sixteen FR residues of 2 other antigen-reactive chicken antibodies, PC17 and VCM2 were randomized with the degenerate codon (NNK) generating 16 sets of scFv library. Amino acids were classified into 5 group of amino acid based on their physicochemical properties: [GAPVLIM], blue; [FYW], purple; [STCHQ], green; [KRH], red; [DE], orange. Positive rate was defined as the percentage of positive clones among 92 clones tested in each set of scFv library. |




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download