INTRODUCTION
MATERIALS AND METHODS
Primary gingival fibroblasts
Cell stimulation
Quantitative real-time polymerase chain reaction (PCR) analysis
Table 1
Primer sequences

IL11 DNA methylation
Statistical analysis
RESULTS
TGF-β1 increased the expression of its target genes with and without 5-aza
 | Figure 1TGF-β1 increased the respective target genes with and without 5-aza. Human gingival fibroblasts were exposed to 5-aza or left untreated for 72 hours, before cells were stimulated with recombinant human TGF-β1. After 24 hours, the gene expression analysis was performed. (A) IL11 (n=10), (B) PRG4 (n=7), (C) NOX4 (n=11); data from individual experiments, denoted with separate colors, indicate the x-fold changes of gene expression in response to TGF-β1 compared to unstimulated cells. P values were obtained from the paired t-test. Statistical analyses were based on fold-change values of single cell donors and experiments. The colors represent data from different cell donors and experiments.
TGF-β1: transforming growth factor-β1, 5-aza: 5-aza-2'-deoxycytidine, IL11: interleukin-11, PRG4: proteoglycan 4, NOX4: NADPH oxidase 4, w/o: without.
|
Table 2
TGF-β1 increased the respective target genes with and without 5-aza

5-aza moderately increased the expression of TGF-β1 target genes
 | Figure 25-aza sensitizes cells to TGF-β1 as indicated by IL11 expression. Human gingival fibroblasts were exposed to 5-aza or left untreated for 72 hours, before cells were stimulated with recombinant human TGF-β1. After 24 hours, gene expression analysis was performed. (A) IL11 (n=10), (B) PRG4 (n=7), (C) NOX4 (n=13), with data from different experiments marked with separate colors. Data indicate the x-fold changes in response to 5-aza in untreated and TGF-β1 treated cells, respectively. P values were obtained from the paired t-test. Statistical analyses were based on fold-change values of single experiments. The colors represent data from different cell donors and experiments.
5-aza: 5-aza-2'-deoxycytidine, TGF-β1: transforming growth factor-β1, IL11: interleukin-11, PRG4: proteoglycan 4, NOX4: NADPH oxidase 4, w/o: without.
|
Table 3
5-aza sensitizes cells to TGF-β1 as indicated by IL11 expression

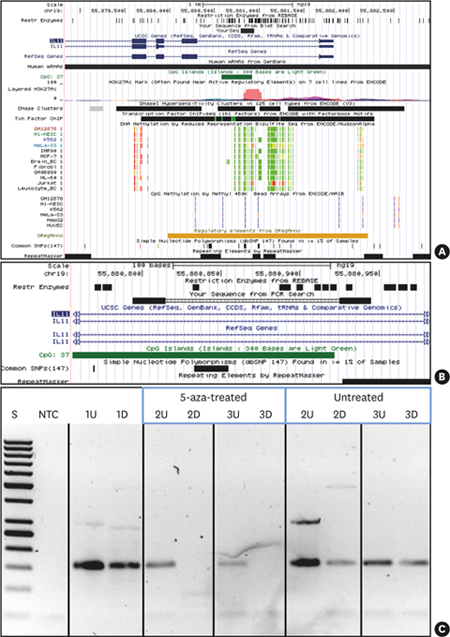
 | Figure 35-aza treatment causes demethylation of IL11 CpG island methylation. UCSC genome browser (hg19) view indicating the location of the IL11 CpG island and PCR amplicon investigated. (A) IL11 gene region (UCSC genome browser view; hg19) presenting the targeted CpG island (intron 1 within IL11: dark green) and the PCR product (indicated as black bar denoted “YourSeq”). DNA methylation tracks (methyl reduced representation bisulfite sequencing; light green indicated 0% and red indicated 100% methylated CpGs). Only Jurkat and HeLa show methylation of the IL11 CpG island. (B) detailed location of the IL11 CpG island and PCR amplicon (primers, bold black) and location of methylation-sensitive restriction sites (denoted as “Rest Enzymes”). (C) IL11-PCR DNA methylation testing results. 1, leukocyte DNA from a normal healthy individual (not 5-aza-treated); 2 and 3, fibroblast DNA: 5-aza-treated vs. non-treated as indicated.
5-aza: 5-aza-2'-deoxycytidine, CpG: commonly next to a guanosine, UCSC: University of California, Santa Cruz, IL11: interleukin-11, PCR: polymerase chain reaction, U: undigested, D: digested by methylation-sensitive restriction enzymes, S: 50 bp ladder, NTC: non-template PCR control 1.
|
5-aza increased the expression of TGF-βRII
Table 4
5-aza increases TGF-βRII in gingival fibroblasts, but not in the presence of TGF-β1

Table 5
TGF-β1 increases TGF-βRI without 5-aza and decreases TGF-βRII in the presence of 5-aza

Table 6
SB431542 blocks TGF-β1 mediated effects on gene expression

5-aza increased the expression of DNMTs: DNMT1 and DNMT3B
Table 7
5-aza increases DNMT1 and DNMT3B, with a similar trend in the presence of TGF-β1





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download