This article has been
cited by other articles in ScienceCentral.
Abstract
The present study investigated prevalence of integrase strand transfer inhibitors (INSTI) resistance mutations in HIV-1-infected antiretroviral therapy (ART)-naïve patients in Korea. From 106 plasma samples, amplification and sequencing of integrase genes was performed, and major or minor mutations were calculated by the Stanford HIV drug resistance mutation interpretation algorithm. No major INSTI resistance mutations were found, and 14 minor mutations were detected in 13 (12.3%) patients. The present data support the recommendation that routine testing for INSTI resistance mutations before starting ART is not necessary.
Keywords: HIV, Integrase Strand Transfer Inhibitors, Mutation, Prevalence, Korean
Integrase strand transfer inhibitors (INSTIs), approved for clinical use in 2007, form the latest class of antiretroviral drugs used to treat human immunodeficiency virus (HIV). Because of their good tolerability and efficacy, INSTIs have become a part of the preferred first-line antiretroviral therapy (ART) regimens in both US and European HIV treatment guidelines.
12 ART regimens that include raltegravir (RAL), a first generation INSTI, have shown long-term efficacy and safety in treatment-naïve and treatment-experienced patients,
345 even in patients with treatment failure due to triple-class drug-resistant HIV.
6 Elvitegravir (EVG) is also widely used as a component of first-line INSTI-based ART; it is formulated as a single-tablet fixed-dose combination together with emtricitabine, tenofovir disoproxil fumarate, and cobicistat. Considering the current widespread use of INSTIs, the emergence of drug resistance mutations is an important issue. Indeed, RAL and EVG have a low genetic barrier to resistance compared with boosted protease inhibitors (PIs). Indeed, data from the period 2009–2012 in the US showed that 15.6% of the 3,012 patients who underwent drug resistance mutation testing had RAL or EVG resistance mutations.
7 However, although RAL or EVG form part of the preferred starting regimen in treatment-naïve HIV-infected patients and the use of this class of antiretroviral drug is increasing, drug resistance mutation testing for INSTIs is not routinely performed before starting ART. This is because the frequency of resistance is low. However, insufficient data exist on the resistance profile of INSTIs in the ART-naïve population worldwide. For these reasons, this study aimed to evaluate of prevalence of and to characterize INSTI resistance mutations among ART-naïve patients in Korea.
The study population included patients aged > 18 years with confirmed HIV-1 infection who were ART-naïve. Patients infected with HIV-2 were excluded. Clinical data, including epidemiologic characteristics, CD4
+ T-cell count, and HIV RNA levels, were collected for all patients via the electronic medical record system. Follow-up data, including CD4 T-cell count and HIV RNA levels, were evaluated 1 year after enrolment. In order to analyze drug resistance, EDTA (ethylenediamine tetraacetic acid) plasma samples from all participating individuals were obtained. Sample preparation, amplification by means of reverse transcription polymerase chain reaction (RT-PCR), and sequencing of the integrase gene were performed using the ViroSeq™ HIV-1 Genotyping Kit (Abbott Laboratories, Abbott Park, IL, USA). Sequenced data were analyzed using ViroSeq™ Integrase Software version 1.0 (Abbott Laboratories). Major and minor integrase resistance mutations were calculated according to the Stanford University HIV Drug Resistance Database genotypic resistance interpretation algorithm.
8 Sequencing and analysis of reverse transcriptase and protease genes were performed by an ABI 3130 analyzer (Applied Biosystems by Thermo Fisher Scientific, Foster City, CA, USA). Collected clinical data were analyzed using the χ
2 test or Fisher's exact test for categorical variables, and the Mann-Whitney test for continuous variables using SPSS version 18.0 for Windows (SPSS Inc., Chicago, IL, USA). A
P value < 0.05 was considered statistically significant. The protocol of this study was approved by the Institutional Review Board of the National Medical Center of Korea (approval No. H-1403/040-005), and written informed consent was obtained from each patient.
In total, 106 HIV-1-infected, ART-naïve patients attending the National Medical Center of Korea were enrolled in this study between March 2014 and August 2015. The majority (99.1%) were male and the median age was 34.0 years (interquartile range [IQR], 27.8–44.0 years). The median CD4 T-cell count at the time of obtaining samples was 292 cells/mm
3 (IQR, 584–1,217 cells/mm
3) and the median HIV-RNA level was 40,712 copies/mL (IQR, 10,282–137,935 copies/mL). The prevalence of resistance mutations is shown in
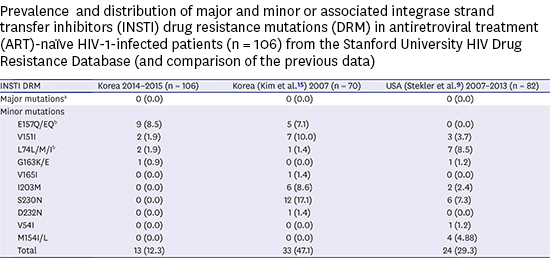
Table 1. No major mutations conferring a marked reduction in viral susceptibility to EVG or RAL were found. However, 14 minor mutations were found in 13 (12.3%) patients: E157Q/EQ was present in 9 (8.5%) samples, L74L/M/I and V151I were each found in 2 (1.9%) samples, G163k was found in 1 (0.94%) sample, and in 1 patient's sample both E157Q and L74M were detected. Regarding reverse transcriptase inhibitor (RTI) and PI resistance mutations, 35.9% of patients had RTI resistance mutations. Sixteen major RTI mutations were determined in 13 (12.6%) patients: V179D was most common (n = 5 [4.9%]), followed by K103N (n = 3 [2.9%]); M41L and T69N (n = 2 [1.9%] each); and V179E, A179D, K238T, and E138K (n = 1 [0.97%] each). Minor RTI resistance mutations were found in 21 patients: V118I (n = 20 [19.4%]) and K103R (n = 1 [0.97%]). No major PI mutations were detected, but minor PI mutations were detected in 51 (49.5%) patients: L10I (n = 39 [37.9%]); L10V (n = 5 [4.9%]); A71V (n = 4 [3.9%]); and V11I, L14V, and V71V (n = 1 [0.97%] each). Of the patients who had INSTI resistance mutations, the most common RTI mutations were V118I (6/13 [46.2%]) and L10I (5/13 [38.5%]) and the most common PI mutation was A71V (3/13 [23.1%]). Factors associated with the presence of INSTI drug resistance mutations, including minor mutations, are shown in
Table 2. Age, sex, initial CD4 T-cell count, initial HIV RNA level, and presence of RTI or PI mutations (including polymorphisms) were not associated with INSTI drug resistance mutations. There were no cases of treatment failure 1 year after starting ART in either group of patients (those with or those without INSTI drug resistance mutations). No significant difference was found in the mean increase in CD4 T-cell count (294 cells/mm
3 vs. 302 cells/mm
3,
P = 0.833) or in the proportion of patients with an HIV RNA level < 40 copies/mL (100% vs. 92.5%,
P > 0.99).
Comparing this study with a similar report conducted in 2007, before the introduction of RAL or EVG in Korea, major mutations were still not identified despite the continued and increasing use of these drugs since 2008 (RAL) and 2014 (EVG). Moreover, no increase in the rate of minor mutations was observed. These findings are similar to those derived from data from the US for the period 2007–2013 (
Table 1).
9 E157Q, a minor mutation detected in our study, is the most common polymorphic mutation selected in patients receiving RAL
10; it is also selected by EVG in vitro. Several studies have shown that E157Q itself has minimal impact on integrase strand transfer activity and viral infectivity.
11 However, E157Q is able to restore damaged enzymatic activity caused by R263K or N155H substitution as a compensatory mutation.
711 G163K is non-polymorphic mutation selected by RAL; it occurs with other INSTI resistance mutations, particularly N155H.
12 V151I, another non-polymorphic mutation, is selected by RAL, while L74M, selected by RAL or EVG, is a relatively common polymorphic accessory INSTI resistance mutation, found in 2.5% of treatment naïve patients,
12 similar to the prevalence found in our data. Although the presence of an L74M mutation alone is not associated with significantly reduced drug susceptibility, in combination with other major drug resistance mutations it could reduce viral susceptibility to INSTIs, including to dolutegravir.
13
In treatment-naïve patients, INSTI-based combination ART is used widely as an initial regimen due to its efficacy and excellent tolerability. Indeed, 93% of enrolled patients were initiated on INSTI-based regimens in our study (data not shown). However, INSTIs such as RAL or EVG have a low genetic barrier to the emergence of resistance. Given the concern of emerging resistance due to the widespread use of INSTIs, several studies have investigated the prevalence of INSTI drug resistance mutations in ART-naïve patients. Between 2006 and 2007, before the introduction of INSTIs in Europe, the European SPREAD HIV resistance surveillance system evaluated the natural genotypic variation of the HIV-1 integrase gene. Of 278 patients tested, no major drug resistance mutations were detected, but only minor resistance mutations were detected in 11 (4%) patients.
14 In the US, from 2007 to 2013, plasma samples were collected from 82 ART-naïve patients; no major drug resistance mutations were found.
9 In Korea, INSTI resistance mutation testing was performed in 75 ART-naïve subjects before the introduction of RAL or EVG. Only minor mutations (including L74M, V151I, and E157Q) were detected; no major mutations were found.
15 As mentioned above, primary resistance due to major mutations remains rare. However, cases of HIV infection with transmitted resistance mutations have begun to emerge. Indeed, several cases of transmitted INSTI drug resistance mutations have been reported.
161718 Furthermore, 2 major INSTI mutations (S147G, N155H) were detected among 840 treatment-naïve patients in the US during the period 2010–2016.
19 In terms of the association between RTI or PI resistance mutations and INSTI resistance mutations, while our study found no significant associations, some integrase polymorphisms found in INSTI-naïve patients were found at an increased frequency in patients with ART failure associated with RTI resistance mutations; hence, such polymorphisms might contribute to the evolution of resistance under INSTI drug pressure.
20 In conclusion, our data support the recommendation that routine testing for integrase gene mutations before starting ART is not necessary. Nevertheless, given the greater use of INSTIs and concerns about the transmission of drug-resistant HIV strains, close monitoring of INSTI resistance is needed.
Figures and Tables
Table 1
Comparison of the distribution of major and minor or associated INSTI DRM in ART-naïve HIV-1-infected patients from studies from the Stanford University HIV Drug Resistance Database

|
INSTI DRM |
Korea 2014–2015 (n = 106) |
Korea (Kim et al.15) 2007 (n = 70) |
USA (Stekler et al.9) 2007–2013 (n = 82) |
|
Major mutationsa
|
0 (0.0) |
0 (0.0) |
0 (0.0) |
|
Minor mutations |
|
|
|
|
E157Q/EQb
|
9 (8.5) |
5 (7.1) |
0 (0.0) |
|
V151I |
2 (1.9) |
7 (10.0) |
3 (3.7) |
|
L74L/M/Ib
|
2 (1.9) |
1 (1.4) |
7 (8.5) |
|
G163K/E |
1 (0.9) |
0 (0.0) |
1 (1.2) |
|
V165I |
0 (0.0) |
1 (1.4) |
0 (0.0) |
|
I203M |
0 (0.0) |
6 (8.6) |
2 (2.4) |
|
S230N |
0 (0.0) |
12 (17.1) |
6 (7.3) |
|
D232N |
0 (0.0) |
1 (1.4) |
0 (0.0) |
|
V54I |
0 (0.0) |
0 (0.0) |
1 (1.2) |
|
M154I/L |
0 (0.0) |
0 (0.0) |
4 (4.88) |
|
Total |
13 (12.3) |
33 (47.1) |
24 (29.3) |
Table 2
Factors associated with the presence of INSTI DRMs in antiretroviral treatment-naïve HIV-1-infected patients

|
Presence of INSTI DRM (including minor mutationsa) |
Present (n = 13) |
Absent (n = 93) |
P value |
|
Sex (male) |
13 (100.0) |
92 (98.9) |
> 0.99 |
|
Age, yr |
31 (25–42) |
34 (28–44) |
0.528 |
|
Initial CD4 T-cell count, cells/mm3
|
349 (112–428) |
292 (181–440) |
0.950 |
|
Initial HIV RNA viral load, copies/mL |
43,020 (3,285–380,815) |
40,712 (11,511–133,525) |
0.751 |
|
Treatment failure within 1 year |
0/11 (0.0) |
0/88 (0.0) |
1.000 |
|
HIV RNA copies < 40 copies/mL after 1 year |
7/7 (100) |
49/53 (92.5) |
1.000 |
|
Increase in CD4 T-cell count after 1 year of ART, cells/mm3
|
294 (149–468) |
302 (192–369) |
0.833 |
|
Presence of RTI mutations (including minor mutations) |
6/13 (46.2) |
26/90 (28.9) |
0.217 |
|
Presence of PI mutations (including minor mutations) |
9/13 (69.2) |
61/90 (67.8) |
1.000 |






 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download