1. Biasutto SN, Caussa LI, Criado del Río LE. Teaching anatomy: cadavers vs. computers? Ann Anat. 2006; 188(2):187–190.

2. Lewis TL, Burnett B, Tunstall RG, Abrahams PH. Complementing anatomy education using three-dimensional anatomy mobile software applications on tablet computers. Clin Anat. 2014; 27(3):313–320.

3. Blume A, Chun W, Kogan D, Kokkevis V, Weber N, Petterson RW, et al. Google bodyBody: 3D human anatomy in the browser. In : Proceeding ACM SIGGRAPH 2011 Talks; 2011 Aug 7–11; Vancouver, Canada. New York, NY: ACM;2011. p. Article no. 19.
4. Shimizu Y, Kawaguchi K, Kanai S. Constructing MRI-based 3D precise human hand models for product ergonomic assessments. In : Proceedings of 2010 Asian Conference on Design and Digital Engineering; 2010 Aug 25–28; Jeju, Korea. Seoul: Society for Computational Design and Engineering;2010. p. 837–844.
5. Petersson H, Sinkvist D, Wang C, Smedby Ö. Web-based interactive 3D visualization as a tool for improved anatomy learning. Anat Sci Educ. 2009; 2(2):61–68.

6. Park JS, Chung MS, Hwang SB, Lee YS, Har DH, Park HS. Visible Korean Human: improved serially sectioned images of the entire body. IEEE Trans Med Imaging. 2005; 24(3):352–360.
7. Park JS, Chung MS, Hwang SB, Lee YS, Har DH. Technical report on semiautomatic segmentation using the Adobe Photoshop. J Digit Imaging. 2005; 18(4):333–343.

8. Shin DS, Park JS, Park HS, Hwang SB, Chung MS. Outlining of the detailed structures in sectioned images from Visible Korean. Surg Radiol Anat. 2012; 34(3):235–247.

9. Shin DS, Chung MS, Park JS, Park HS, Lee S, Moon YL, et al. Portable document format file showing the surface models of cadaver whole body. J Korean Med Sci. 2012; 27(8):849–856.

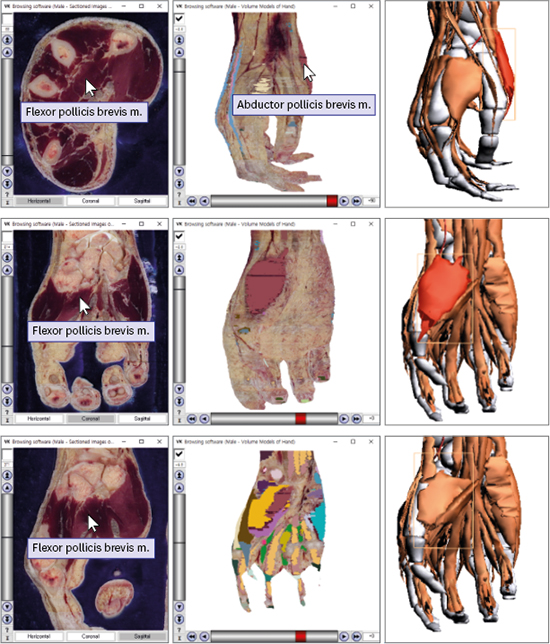
10. Shin DS, Kwon K, Shin BS, Park HS, Lee S, Lee SB, et al. Surface models and gradually stripped volume model to explore the foot muscles. Anatomy. 2015; 9(1):19–25.

11. Chung BS, Kwon K, Shin BS, Chung MS. Surface models and gradually peeled volume model to explore hand structures. Ann Anat. 2017; 211:202–206.

12. Shin DS, Chung MS, Park HS, Park JS, Hwang SB. Browsing software of the Visible Korean data used for teaching sectional anatomy. Anat Sci Educ. 2011; 4(6):327–332.

13. Kwon K, Chung MS, Park JS, Shin BS, Chung BS. Improved software to browse the serial medical images for learning. J Korean Med Sci. 2017; 32(7):1195–1201.

14. Shin DS, Chung MS, Shin BS, Kwon K. Laparoscopic and endoscopic exploration of the ascending colon wall based on a cadaver sectioned images. Anat Sci Int. 2014; 89(1):21–27.

15. Kwon K, Shin DS, Shin BS, Park HS, Lee S, Jang HG, et al. Virtual endoscopic and laparoscopic exploration of stomach wall based on a cadaver’s sectioned images. J Korean Med Sci. 2015; 30(5):658–661.

16. Shin DS, Lee S, Park HS, Lee SB, Chung MS. Segmentation and surface reconstruction of a cadaver heart on Mimics software. Folia Morphol (Warsz). 2015; 74(3):372–377.

17. Moore KL, Dalley AF, Agur AM. Clinically Oriented Anatomy. 7th ed. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health;2014.




 PDF
PDF Citation
Citation Print
Print



 XML Download
XML Download