Abstract
Purpose
To investigate the effect and mechanism of phospholipase C epsilon gene 1 (PLCE1) expression on esophageal cancer cell lines.
Materials and Methods
The esophageal carcinoma cell lines Eca109 and EC9706 and normal esophageal epithelial cell line HEEC were cultured. The expression of PLCE1, protein kinase C alpha (PKCα), and nuclear factor kappa B (NF-κB) p50/p65 homodimer in cells were comparatively analyzed. The esophageal cancer cells were divided into si-PLCE1, control siRNA (scramble), and mock groups that were transfected with specific siRNA for PLCE1, control siRNA, and blank controls, respectively. Expression of PLCE1, PKCα, p50, and p65 was detected by Western blotting. Transwell assay was used to detect migration and invasion of Eca109 and EC9706 cells.
Results
Compared with HEEC, the expression of PLCE1, PKCα, p50, and p65 was increased in Eca109 and EC9706 cells. The expression of PLCE1 was positively correlated with the expression of PKCα and p50 (PKCα: r=0.6328, p=0.032; p50: r=0.6754, p=0.041). PKCα expression had a positive correlation with the expression of p50 and p65 (p50: r=0.9127, p=0.000; p65: r=0.9256, p=0.000). Down-regulation of PLCE1 significantly decreased the expression of PKCα and NF-κB-related proteins (p65: p=0.002, p=0.004; p50: p=0.005, p=0.009) and inhibited the migration and invasion of Eca109 and EC9706 cells.
Esophageal carcinoma is a malignant tumor with high incidence worldwide and is the fourth cause of cancer death in China.1 The occurrence of esophageal cancer has been shown to be closely related to inflammation, in addition to smoking, dietary habits, and genetic polymorphism.2 The pathogenesis of esophageal cancer, especially in regards to the molecular mechanism thereof, has recently drawn great interest.
The gene phospholipase C epsilon 1 (PLCE1) has been shown to be associated with susceptibility to esophageal34 and other cancers. Wang, et al.5 determined that the expression of PLCE1 could be used to diagnose and predict the stage of colorectal cancer. Meanwhile, others found that the growth of bladder cancer cells is inhibited upon transfection with si-PLCE1.6 Chen, et al.7 noted a positive correlation between PLCE1 and gastric cancer occurrence.
Previous studies have proven that the occurrence of esophageal cancer is closely related to inflammation8 and that nuclear factor kappa B (NF-κB) is a key factor in inflammation-related tumors.9 High expression of NF-κB related proteins in esophageal cancer suggests that NF-κB signaling may be activated in esophageal cancer.10 Interestingly, Zhao, et al.11 found that protein kinase C (PKC) could activate multiple signaling pathways, including NF-κB and that it could be activated indirectly by phospholipase C: PKC plays an important role in cell proliferation, differentiation, apoptosis, hormone secretion, and gene expression, and PKC is a key link in a series of cell signal transduction pathways.12
In light of the studies above, we speculated that PLCE1, PKC alpha (PKCα), and NF-κB may indirectly activate the development of esophageal cancer and promote the progression of esophageal cancer. Accordingly, we aimed to investigate and compare the expression of PLCE1, PKCα, and NF-κB-related proteins in esophageal cancer cells (Eca109, EC9706) and normal esophageal epithelial cell line HEEC. We also analyzed correlations among the expressions of PLCE1, PKCα, and NF-κB-related protein to outline the mechanism and impact of PLCE1 expression in esophageal cancer.
All cell lines were purchased from the Shanghai Cell Bank (Shanghai, China). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Thermo Fisher Scientific, Berkeley, SF, USA) containing 10% fetal bovine serum, 2 mM glutamine, 100 kU/L penicillin, and 0.1% streptomycin and incubated in a humidified chamber at 37℃ with 5% CO2.
PLCE1 siRNA (sc-44024) and control siRNA (sc-37007) were purchased from Santa Cruz Biotech (Santa Cruz, CA, USA). Sc-44024 and sc-37007 were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) into Eca109 and EC9706 cells in the logarithmic phase of growth. The cells were divided into a si-PLCE1, scramble, or mock group. All transfection operations were carried out strictly according to the manufacturer's instructions. Then the cells were incubated in a humidified chamber at 37℃ with 5% CO2.
Specimens were surgically obtained from 30 consecutive patients (18 men and 12 women) with esophageal squamous cell carcinoma from November 2016 to December 2017. The mean age was 59.1 years (range, 38–77 years). None of them received irradiation or chemotherapy preoperatively. The representative cancerous lesions including adjacent noncancerous mucosa were taken.
RNA was isolated from dissected ocular tissues using commercial kits based on guanidinium isothiocyanate extraction methods (ToTALLY RNA, Ambion, Austin, TX, USA). For each case, small amounts of tissue (<50 mg) were homogenized in 1 mL of lysis solution using a motorized rotor-stator homogenizer (Brinkmann Instruments, Inc., Westbury, NY, USA). To prevent filter clogging during silica-based filter binding, the homogenate was repeatedly centrifuged to remove debris and insoluble material before proceeding to filtration. Further extraction steps were performed according to the manufacturer's protocols. RNA quality was assessed by agarose gel electrophoresis.
Total RNA was extracted using TRIzol and quantified. Then cDNA was reverse-transcribed and quantified by PrimeScript RT reagent kits (Takara, Tokyo, Japan) and TaqMan MicroRNA assay kits (Applied Biosystems, Foster City, CA, USA), respectively. β-actin was used as a reference gene for PLCE1 and PKCα. The primer sequences were as follows: PLCE1 F: 5′-GAGCTG CAATCGAAGTCTGG-3′; R: 5′-AAGGCCTTCTGTGAGTCCTC-3′. PKCα F: 5′-GCTAGCTGGGCAGCTTAT-GA-3′; R: 5′-CCAGCT GATCTGGTGGTGTT-3′. β-actin F: 5′-CTCCATCCTGGC-CTC GCTGT-3′; R: 5′-GCTGTCACCTTCACCGTTCC-3′. The relative gene expression was analyzed with the 2−ΔΔCt method.
Total protein was extracted from cells and determined by BCA Protein Assay Kits (Pierce, Shanghai, China). Subsequently, 50 µg proteins were subjected to SDS-PAGE electrophoresis and transferred onto membranes. The membranes were incubated with PLCE1 antibody (1:1000, DF2565, Affinity, Cincinnati, OH, USA), PKCα antibody (1:1000, ab31, Abcam, Cambridge, UK), p50 antibody (1:1000, PA1-30409, Invitrogen), or p65 antibody (1:1000, acetyl K310, Abcam) overnight at 4℃ separately. Thereafter, horseradish peroxidase-labeled secondary antibody (1:5000, Beijing Zhong Shan Biotechnology Co., Ltd., Beijing, China) was added for 1 h. Proteins were visualized with an enhanced chemiluminescence kit and gel imaging system. The experiments were repeated three times.
1×104 cells were seeded in 96 well plates. Cells were divided into si-PLCE1, mock, and scramble groups, and then, they continued to culture for 72 h. For Transwell migration assays, 1×105 cells were plated in the top chamber with the non-coated membrane (pore size, 8 mm; BD Biosciences, Franklin Lake, NJ, USA). For invasion experiments, cells were plated in the top chamber with 1 mg/mL Matrigel-coated membrane (pore size, 8 mm; BD Biosciences) for 1 h. Cells were plated in serum-free medium, and DMEM medium containing 5% fetal bovine serum was added in the lower chamber. After incubation for 24 h, the cells in the top chamber were removed. The cells on the lower surface of membrane were fixed in 4% polyformaldehyde solution for 15 min. Then, the cells were stained with 0.05% crystal violet for 40 min and counted under a Leica DC 300F microscope (Oskar-Barnack, Solms, Germany).
All data are represented by mean values±standard deviation and were analyzed with SPSS version 21.0 (IBM Corp., Armonk, NY, USA). LSD-t test was applied to analyze differences between two groups. Correlation analysis was analyzed by Spearman's rank correlation, and p<0.05 was considered as statistically significant.
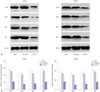
The expression levels of PLCE1 and PKCα in Eca109, EC9706, and HEEC were detected by qRT-PCR and Western blotting. The expression of PLCE1 in Eca109 and EC9706 cells was higher than that in HEEC (p=0.002, p=0.003) (Fig. 1A and B). Similarly, the expression of PKCα was significantly increased in Eca109 and EC9706 (p=0.005, p=0.007), compared to HEEC (Fig. 1C and D).
The expression of NF-κB p50/p65 homodimer was detected by Western blotting. Compared with HEEC group, p50 and p65 expression was increased in Eca109 and EC9706 cells (p65: p=0.002, p=0.004; p50: p=0.005, p=0.009). This indicated that the NF-κB signaling pathway is activated in esophageal cancer cells (Fig. 2).
In this study, Spearman's rank correlation was used to analyze correlations among PLCE1, PKCα, p50, and p65. qRT-PCR analysis results showed that overexpression of PKCα is detected in esophageal cancer tissues, along with high expression of PLCE1, and that the expression of PLCE1 is positively correlated with PKCα expression (r=0.6328, p=0.032). Overexpression of p50 was detected in esophageal cancer tissues, along with high PLCE1 expression, indicating a positive correlation between p50 and PLCE1 (r=0.6754, p=0.041) (Fig. 3A). Further statistical analysis showed that the expression of PKCα in esophageal cancer is positively correlated with the expression of p50 and p65 (r=0.9127, r=0.9256) (Fig. 3B). This suggested that PKCα and NF-κB signaling play a synergistic role in esophageal cancer.
Reportedly, the NF-κB signaling pathway is indirectly activated by PKCα.13 Accordingly, we transfected PLCE1 siRNA into Eca109 and EC9706 cells and further studied whether PLCE1 activates NF-κB signaling by activating PKCα. After transfection, the expression of PLCE1 in the transfected cells was significantly lower (p=0.015). Compared with the mock and scramble groups, the expression levels of PKCα, p50, and p65 protein were lower in the si-PLCE1 group (Eca109: p=0.005, 0.002, 0.001; EC9706: p=0.003, 0.003, 0.006). From this, it could be inferred that PLCE1 activates the NF-κB signaling pathway through PKCα (Fig. 4).
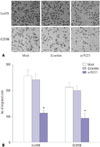
Migration of Eca109 and EC9706 cells was detected by Transwell experiments. Therein, we noted no statistical differences in cell migration ability between the mock and scramble groups. Compared with the mock and scramble groups, however, the migration rate of the si-PLCE1 group was decreased significantly (Eca109: p=0.001; EC9706: p=0.002) (Fig. 5).
Therefore, PLCE1 could promote the migration of esophageal cancer cells by activating NF-κB signaling.
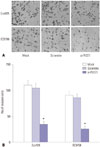
Transwell invasion results also showed that there was no statistical difference in cell invasion ability between the mock group and scramble group. Compared with these two groups, however, the number of Eca109 and EC9706 cells in the si-PLCE1 group passing through the membrane was reduced significantly (Eca109: p=0.000; EC9706: p=0.001) (Fig. 6). This indicated that PLCE1 could promote invasion of esophageal cancer cells by activating NF-κB signaling.
In this study, we found that the expression levels of PLCE1, PKCα, p50, and p65 were increased in Eca109 and EC9706 cells, compared to HEEC. The expression of PLCE1 protein was positively correlated with the expression of PKCα, p50, and p65, and we discovered a positive correlation between the expression of PKCα and p50/p65. Downregulation of PLCE1 decreased the expression of PKCα and NF-κB-related proteins and inhibited the migration and invasion abilities of Eca109 and EC9706 cells.
While PLCE1 expression and related mechanisms in esophageal cancer have been studied, there has been no research on PLCE1 and cell migration and invasion. Studies have shown that gene mutation, allele high frequency heterozygosity deletion, and tumor suppressor gene inactivation could promote the development of esophageal cancer. Moreover, current research has shown that mutation of PLCE1 might promote the development of cancer. Furthermore, two genome-wide association studies on large samples have identified a new esophageal cancer susceptibility locus, rs2274223, on PLCE1 on chromosome 10q23. Gu, et al.14 found that PLCE1 rs2274223 could be used to predict the risk of esophageal cancer. Also, Guo, et al.15 revealed that PLCE1 rs2274223 polymorphism is associated with esophageal cancer. Study has also shown that PLCE1 regulates cell growth, proliferation, and differentiation and affects cytoskeleton change, cell movement, cell apoptosis, tumor growth and development, and other biological behaviors.16 Li, et al.17 demonstrated that PLCE1 is expressed at high levels in esophageal cancer. Zhai, et al.18 indicated that PLCE1 could promote esophageal cancer cell progression through maintaining the transcriptional activity of Snail. In addition, Zhao, et al.19 found that the expression of PLCE1 was increased and inhibited apoptosis in esophageal carcinoma cells.
Consistent with previous studies, the expression of PLCE1 was upregulated in this study. In addition, low expression of PLCE1 could inhibit cell migration and invasion and decrease the expression of PKCα and NF-κB-related proteins. Another study found that PLCE1 plays an important role in cell signal transduction.20 NF-κB signaling is involved in cell migration and invasion21 via two subunits (p50 and p65).22 Ordoñez-moreno, et al.23 revealed that overexpression of NF-κB p50/p65 homodimer promotes migration and invasion in mammary non-tumorigenic epithelial cells. Meanwhile, TGF-β1 can promote migration and invasion of Human Squamous Cervical Carcinoma Line Siha cells by upregulating the expression of p50 and p65.24 Additionally, Cui, et al.25 found that the expression of PLCE1 is increased in Kazakh patients with esophageal carcinoma and is strongly correlated with NF-κB signaling. The above studies illustrated that PLCE1 promotes the invasion and migration of esophageal cancer cells through the NF-κB signaling pathway. Activation of PKC proteins is involved in signal transduction pathways, although it usually exists in on-activated cytoplasm, and the subunit of PKC is a sign of activation.26 PKCα is one of the subunits of PKC, and it plays an important role in the transduction from the cytoplasm to the nucleus.27 Cui, et al.25 also showed that PKCα is increased in esophageal cancer tissues. In our study, down-regulation of PLCE1 decreased the expression of PKCα in esophageal cancer cells. This indicated that PLCE1 could regulate the expression of PKCα. Similarly, Zhao, et al.11 discovered that Sesamin had a certain inhibitory effect on mast cell activation by inhibiting PKCα/NF-κB signaling. Also, high density lipoprotein promoted the inflammatory effect of macrophages through PKCα/NF-κB signaling.28 Accordingly, we discerned that PLCE1 is involved in esophageal cancer via the PKCα/NF-κB pathway.
There are some limitations in this study. Due to limitations of experimental conditions, only silencing PLCE1 was applied in this study to test our hypothesis of PLCE1 activating NF-kB through PKCa, and additional experiments will be needed to show how PLCE1 modulates the NF-kB pathway. In addition, it is not clear whether NF-kB signaling is governed by PLCE1, and the synergistic effect of inhibition of PLCE1 and NF-kB appears to be the summation of inhibition of both pathways, rather than evidence of a downstream effect of PLCE1 to NF-kB. The above problems will be confirmed in future experiments.
In brief, PLCE1 appears to activate the NF-κB signaling pathway by regulating PKCα to promote the invasion and migration of esophageal cancer cells. This may become a new target for the treatment of esophageal cancer.
Figures and Tables
Fig. 1
Expression of PLCE1 and PKCα in esophageal cancer cells. (A) qRT-PCR was used to detect the expression of PLCE1 mRNA in esophageal cancer cells. (B) The expression of PLCE1 was detected by Western blotting. (C) The expression of PKCα mRNA. (D) The expression of PKCα protein and its statistical analysis map. Compared with HEEC, *p<0.01.

Fig. 2
Expression of NF-κB signaling pathway-related proteins. (A) Western blotting was used to detect the expression of p50 and p65. (B) Statistical analysis of P50 and p65 expression. Compared with HEEC, *p<0.01.

Fig. 3
Correlation analysis of PLCE1, PKCα, and p50/p65 protein expression in esophageal cancer cells. (A) Relationships among PLCE1, PKCα, and p50/p65. (B) Relationship between PKCα and p50/p65.

Fig. 4
Effect of PLCE1 on PKCα and p50/p65 expression in esophageal cancer cells. (A) After transfection with PLCE1 siRNA, the expression of PLCE1, PKCα, and p50/p65 was detected by Western blotting. (B) Statistical analysis map of PLCE1, PKCα, and p50/p65. Compared with the mock group or scramble group, *p<0.05, †p<0.01.
References
1. Yang Q, Lin W, Liu Z, Zhu J, Huang N, Cui Z, et al. RAP80 is an independent prognosis biomarker for the outcome of patients with esophageal squamous cell carcinoma. Cell Death Dis. 2018; 9:146.

2. Zhang M, Zhang L, Cui M, Ye W, Zhang P, Zhou S, et al. miR-302b inhibits cancer-related inflammation by targeting ERBB4, IRF2 and CXCR4 in esophageal cancer. Oncotarget. 2017; 8:49053–49063.

3. Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010; 42:759–763.
4. Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO, et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010; 42:764–767.

5. Wang ZJ, Wang DX, Wang ZG, Pathology DO, Hospital ZP, Pediatrics DO. Expression of PLCE1 and p53 in colorectal cancer and its significance. J Clin Exp Med. 2018; 3:247–251.
6. Cheng H, Luo C, Wu X, Zhang Y, He Y, Wu Q, et al. shRNA targeting PLCε inhibits bladder cancer cell growth in vitro and in vivo. Urology. 2011; 78:474.e7–474.e11.
7. Chen H, Zhao J, Ji F. The progress of correlation research about PLCE1 gene and gastric cancer occurrence. J Modern Oncol. 2016; 24:2160–2162.
8. Jomrich G, Paireder M, Gleiss A, Kristo I, Harpain L, Schoppmann SF. Comparison of inflammation-based prognostic scores in a cohort of patients with resectable esophageal cancer. Gastroenterol Res Pract. 2017; 2017:1678584.

9. Lu Z, Long Y, Cun X, Wang X, Li J, Mei L, et al. A size-shrinkable nanoparticle-based combined anti-tumor and anti-inflammatory strategy for enhanced cancer therapy. Nanoscale. 2018; 10:9957–9970.

10. Long L, Pang XX, Lei F, Zhang JS, Wang W, Liao LD, et al. SLC52A3 expression is activated by NF-κB p65/Rel-B and serves as a prognostic biomarker in esophageal cancer. Cell Mol Life Sci. 2018; 75:2643–2661.

11. Zhao HW, Cui YF, Jiang JZ, Che N, Jing YE, Wang CY, et al. [Sesamin suppresses mast cell activation through inhibition of PKCα/NF-κB signaling pathway]. Zhongguo Mian Yi Xue Za Zhi. 2018; 2:167–171.
12. Racchi M, Buoso E, Ronfani M, Serafini MM, Galasso M, Lanni C, et al. Role of hormones in the regulation of RACK1 expression as a signaling checkpoint in immunosenescence. Int J Mol Sci. 2017; 18:1453.

13. Song T, Zheng YM, Vincent PA, Cai D, Rosenberg P, Wang YX. Canonical transient receptor potential 3 channels activate NF-κB to mediate allergic airway disease via PKC-α/IκB-α and calcineurin/IκB-β pathways. FASEB J. 2016; 30:214–229.

14. Gu H, Ding G, Zhang W, Liu C, Chen Y, Chen S, et al. Replication study of PLCE1 and C20orf54 polymorphism and risk of esophageal cancer in a Chinese population. Mol Biol Rep. 2012; 39:9105–9111.

15. Guo LY, Yang N, Hu D, Zhao X, Feng B, Zhang Y, et al. PLCE1 rs2274223 polymorphism and susceptibility to esophageal cancer: a meta-analysis. Asian Pac J Cancer Prev. 2014; 15:9107–9112.

16. Cui XB, Li S, Li TT, Peng H, Jin TT, Zhang SM, et al. Targeting oncogenic PLCE1 by miR-145 impairs tumor proliferation and metastasis of esophageal squamous cell carcinoma. Oncotarget. 2016; 7:1777–1795.

17. Li Y, An J, Huang S, Liao H, Weng Y, Cai S, et al. PLCE1 suppresses p53 expression in esophageal cancer cells. Cancer Invest. 2014; 32:236–240.

18. Zhai S, Liu C, Zhang L, Zhu J, Guo J, Zhang J, et al. PLCE1 promotes esophageal cancer cell progression by maintaining the transcriptional activity of snail. Neoplasia. 2017; 19:154–164.

19. Zhao L, Wei ZB, Yang CQ, Chen JJ, Li D, Ji AF, et al. Effects of PLCE1 gene silencing by RNA interference on cell cycling and apoptosis in esophageal carcinoma cells. Asian Pac J Cancer Prev. 2014; 15:5437–5442.

20. Zhang B, Liu T, Zhou H. [The promoting research of phospholipase C epsilon-1 on nasal Th2 cell polarization]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2014; 28:1363–1366.
21. Li J, Li Y, Wang B, Ma Y, Chen P. Id-1 promotes migration and invasion of non-small cell lung cancer cells through activating NF-κB signaling pathway. J Biomed Sci. 2017; 24:95.

22. Che R, Zhang A. Mechanisms of glucocorticoid resistance in idiopathic nephrotic syndrome. Kidney Blood Press Res. 2013; 37:360–378.

23. Ordoñez-Moreno A, Rodriguez-Monterrosas C, Cortes-Reynosa P, Perez-Carreon JI, Perez Salazar E. Erythropoietin induces an epithelial to mesenchymal transition-like process in mammary epithelial cells MCF10A. J Cell Biochem. 2017; 118:2983–2992.

24. Mao M, Cai L, Hu J, Hu L. Investigating the migration and invasion of human squamous cervical carcinoma line Siha stimulated by TGF-β1 and its underlying mechanisms. Chin J Cell Biol. 2015; 1:59–65.
25. Cui XB, Pang XL, Li S, Jin J, Hu JM, Yang L, et al. Elevated expression patterns and tight correlation of the PLCE1 and NF-κB signaling in Kazakh patients with esophageal carcinoma. Med Oncol. 2014; 31:791.

26. Hsieh YH, Wu TT, Tsai JH, Huang CY, Hsieh YS, Liu JY. PKCalpha expression regulated by Elk-1 and MZF-1 in human HCC cells. Biochem Biophys Res Commun. 2006; 339:217–225.
27. Lahn M, Paterson BM, Sundell K, Ma D. The role of protein kinase C-alpha (PKC-alpha) in malignancies of the gastrointestinal tract. Eur J Cancer. 2004; 40:10–20.

28. van der Vorst EPC, Theodorou K, Wu Y, Hoeksema MA, Goossens P, Bursill CA, et al. High-density lipoproteins exert pro-inflammatory effects on macrophages via passive cholesterol depletion and PKC-NF-κB/STAT1-IRF1 signaling. Cell Metab. 2017; 25:197–207.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download