Dear Editor,
Pallister-Killian syndrome (PKS; OMIM#601803) is characterized by specific craniofacial dysmorphism, pigmentary skin anomalies, limb differences, congenital heart defects, hypotonia, intellectual disabilities, and epilepsy. PKS is caused by the presence of extra copies of the short arm of chromosome 12, which most commonly presents as supernumerary marker isochromosome 12p [i(12p)], resulting in 12p tetrasomy [1].
The critical region for PKS is located at 12p13.3, and the strongest candidate genes for the PKS phenotype are ING4 and CHD4. ING4 belongs to the inhibitor of growth family and plays important roles in transcriptional regulation. Overexpression of ING4 negatively regulates cell growth, resulting in cell cycle arrest and apoptosis. CHD4 is a chromodomain helicase DNA binding protein, which plays an important role in chromatin remodeling [2].
Supernumerary markers are often in a mosaic state; they can be present solely in some tissues and absent in others, and the mosaic ratio can vary quite significantly. i(12p) is reported to be unstable with age and is less frequent in highly replicative tissues, likely because it is lost with cell division [3]. Classic cytogenetic techniques can fail to identify i(12p), even if the clinical features are very suggestive of the syndrome. Peripheral blood lymphocytes, routinely used for chromosome diagnosis, are less suitable for analysis since they usually show a low frequency of this supernumerary chromosome; moreover, they have a high replication rate and have to be cultured [4]. Phytohemagglutinin, used to stimulate lymphocyte division, likely promotes the growth of normal euploid cells, resulting in underrepresentation of i(12p) [5]. Fibroblasts are usually more informative since they contain a higher proportion of i(12p) cells; however, metaphases are obtained after cell culture, which can bias the true mosaic ratio [6]. Moreover, fibroblast analysis requires a skin biopsy, an invasive procedure that can cause the patient higher levels of discomfort.
The array comparative genomic hybridization (CGH) technique can be performed on DNA directly extracted from tissues without cell culture, providing a better representation of the mosaic ratio. A limitation of this technique is its sensitivity; mosaics <20–30% are not detected. i(12p) diminishes over time in peripheral blood, which can be a problem since PKS diagnosis is rarely accomplished during the first year of life [78]. We present a case of PKS diagnosed by array CGH using DNA directly extracted from a buccal swab, where the array profile from peripheral blood DNA was normal. The patient, a one-year-old-boy, was admitted to the Section of Medical Genetics of the Azienda Ospedaliero Universitaria Pisana, Pisa, Italy, in August 2017.
This genetic study was performed with written informed consent obtained from the parents. This study did not require Institutional Review Board approval. Buccal cells were chosen since they are an easy source of DNA when patient compliance is poor; moreover, a cell culture is not required, and the extraction technique is relatively simple. DNA was extracted using QIAamp spin columns (Qiagen, Hilden, Germany), a fast procedure where nucleic acids selectively adsorb on a silica support in the presence of a high concentration of chaotropic salts, while proteins and contaminants are washed away. Although swab techniques have been reported to provide a lower quantity and quality of DNA than whole saliva, the DNA obtained in this study had a high molecular weight and minimal bacterial DNA. We obtained 50 µL of DNA (concentration: 97 ng/µL; 260/280 absorbance ratio: 1.75; 260/230 absorbance ratio: 1.85). This DNA was sufficient for an excellent quality experiment, as indicated by the array CGH analysis software Cytogenomics (Agilent, Santa Clara, CA, USA).
A comparison of the array results from peripheral blood and the buccal swab showed that i(12p) was not detected in the first case, as the chromosome 12 profile was normal, whereas the duplication was evident in the experiment using buccal cell DNA (Fig. 1A). Quality control reports for both experiments were excellent, so differences in results are unlikely to have been influenced by experiment quality.
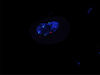
In a single experiment, array CGH provided insights into the extent of duplication, gene content, and mosaic ratio. The probes showed a 34 Mb gain from the probe in position 230,421 (hg19 map, 12p13.33) to the probe in position 34,345,585 (12p11.1), indicating that the entire p arm is present in i(12p), as shown in Fig. 1A. In addition, both ING4 and CHD4 are included in this region. The mean log2 ratio of the 12p probes (0.634) provides some indication regarding the percentage of tetrasomic cells. We next plotted the expected fluorescent value according to the mosaic ratio (Fig. 1A and 1B); in this case, it is predicted that 50–60% of the cells contain i(12p). To validate this result, we performed a FISH test on buccal mucosa nuclei, using a commercial probe (LSI ETV6(TEL)/RUNX1(AML1) probe; Abbott, Abbott Park, IL, USA). Four signals were detected in 46/100 nuclei scored, whereas, as expected, the control probe showed only two signals, confirming the presence of four copies of ETV6 (Fig. 2).
In conclusion, although this is a single case and analyses of additional patients are needed, we have shown that array CGH performed on buccal swab DNA is an easy and sensitive technique for detecting i(12p) that can be used as an initial test to diagnose PKS. Array CGH has many advantages over other techniques such as FISH or droplet PCR [9]. A single experiment provides not only an indication of the mosaic ratio but also precise information regarding the genetic composition of the isochromosome.
Figures and Tables
 | Fig. 1Results of array CGH. (A) Array CGH profile of chromosome 12 performed on (a) peripheral blood DNA and (b) buccal swab where the average log2 ratio is approximately 0.634, consistent with a mosaicism of 50–60% i(12p) cells. (c) Relationship between the percentage of mosaicism (X-axis) and the log2 ratio (Y-axis) of the array CGH experiment is shown. (B) Diagram presenting the relationship between the percentage of i(12p) cells and the log2 ratio obtained. (a) If all the cells in the sample are tetrasomic for the 12p region, the number of copies of i(12p) per cell is 4. The ratio with a normal sample is 4/2=2, with a log2 of 1. (b) If 80% of the cells in the sample are tetrasomic for the 12p region, the number of copies of i(12p) per cell is 3.6. The ratio with a normal sample is 3.6/2=1.8, with a log2 of 0.85. (c and d) The values obtained in cases of 50% and 20% mosaicism are also shown.Abbreviation: CGH, Comparative Genomic Hybridization.
|
References
2. Izumi K, Conlin LK, Berrodin D, Fincher C, Wilkens A, Haldeman-Englert C, et al. Duplication 12p and Pallister-Killian syndrome: a case report and review of the literature toward defining a Pallister-Killian syndrome minimal critical region. Am J Med Genet A. 2012; 158A:3033–3045.


3. Tang W, Wenger SL. Cell death as a possible mechanism for tissue limited mosaicism in Pallister-Killian syndrome. J Assoc Genet Technol. 2005; 31:168–169.
4. Peltomäki P, Knuutila S, Ritvanen A, Kaitila I, de la Chapelle A. Pallister-Killian syndrome: cytogenetic and molecular studies. Clin Genet. 1987; 31:399–405.


5. Reeser SL, Wenger SL. Failure of PHA-stimulated i(12p) lymphocytes to divide in Pallister-Killian syndrome. Am J Med Genet. 1992; 42:815–819.


6. Priest JH, Rust JM, Fernhoff PM. Tissue specificity and stability of mosaicism in Pallister-Killian +i(12p) syndrome: relevance for prenatal diagnosis. Am J Med Genet. 1992; 42:820–824.


7. Lee MN, Lee J, Yu HJ, Lee J, Kim SH. Using array-based comparative genomic hybridization to diagnose Pallister-Killian syndrome. Ann Lab Med. 2017; 37:66–70.






 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download