INTRODUCTION
METHODS
Patient and control selection
Table 1
Results of C-FCM and HS-FCM in 23 patients with aplastic anemia/myelodysplastic syndrome and paroxysmal nocturnal hemoglobinuria
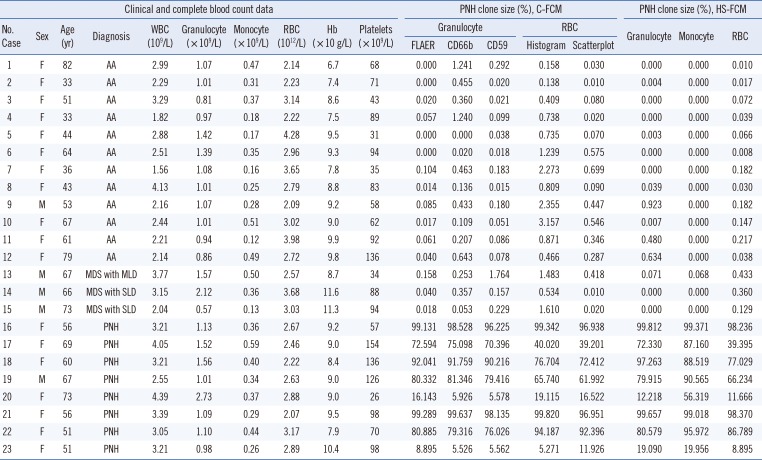
Abbreviations: M, male; F, female; WBC, white blood cell; RBC, red blood cell; Hb, hemoglobin; C-FCM, conventional flow cytometry; HS-FCM, high sensitivity flow cytometry; FLAER, fluorescein-labeled proaerolysin; PNH, paroxysmal nocturnal hemoglobinuria; MDS, myelodysplastic syndrome; SLD, single lineage dysplasia; MLD, multilineage dysplasia; AA, aplastic anemia; CD, cluster of differentiations.
FCM
PNH clone incidence and size in healthy controls
Statistical analysis
RESULTS
LOD and sensitivity of HS-FCM
PNH clone incidence and size in healthy controls
The median acquisition events of C-FCM and HS-FCM in patient samples
Comparison of HS-FCM results obtained from three cell lineages
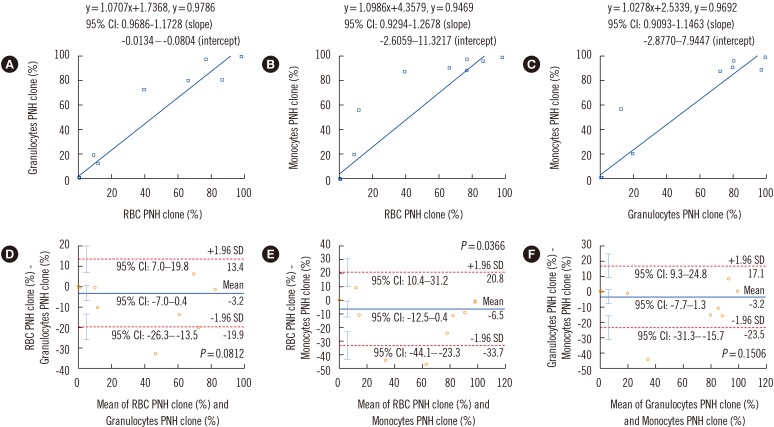 | Fig. 1Comparison of high sensitivity flow cytometry results obtained from three cell lineages (granulocytes, monocytes, and RBCs). Results are compared using Passing-Bablok regression analysis (A-C) and Bland-Altman analysis (D-F). Regression equations, correlation coefficients, and 95% CIs for the slope and intercept of the equations are shown in A-C. In the Bland-Altman analysis, mean difference (indicated by the blue horizontal line), lower and upper limits of agreement (indicated by the red dotted horizontal lines), and their 95% CIs (indicated by the cyan vertical bars) are presented in D-F.Abbreviations: RBC, red blood cell; PNH, paroxysmal nocturnal hemoglobinuria; CI, confidence interval.
|
Comparison of C-FCM and HS-FCM results for granulocytes
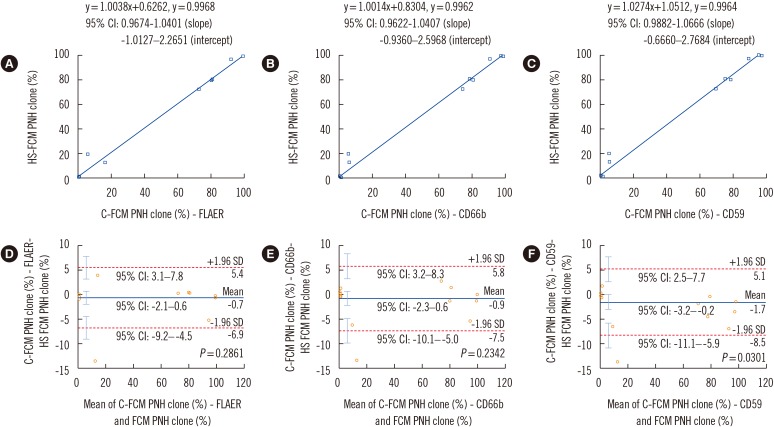 | Fig. 2Comparison of C-FCM granulocyte PNH clone size results with three different GPI markers and HS-FCM. Results are compared using Passing-Bablok regression analysis (A-C) and Bland-Altman analysis (D-F). Regression equations, correlation coefficients, and 95% CIs for the slope and intercept of the equations are shown in (A-C). In the Bland-Altman analysis, mean difference (indicated by the blue horizontal line), lower and upper limits of agreement (indicated by the red dotted horizontal lines), and their 95% CIs (indicated by the cyan vertical bars) are presented in (D-F).Abbreviations: HS-FCM, high sensitivity flow cytometry; C-FCM, conventional flow cytometry; PNH, paroxysmal nocturnal hemoglobinuria; FLAER, fluorescein-labeled proaerolysin; CD, cluster of differentiations; CI, confidence interval.
|
Comparison of C-FCM and HS-FCM results for RBCs
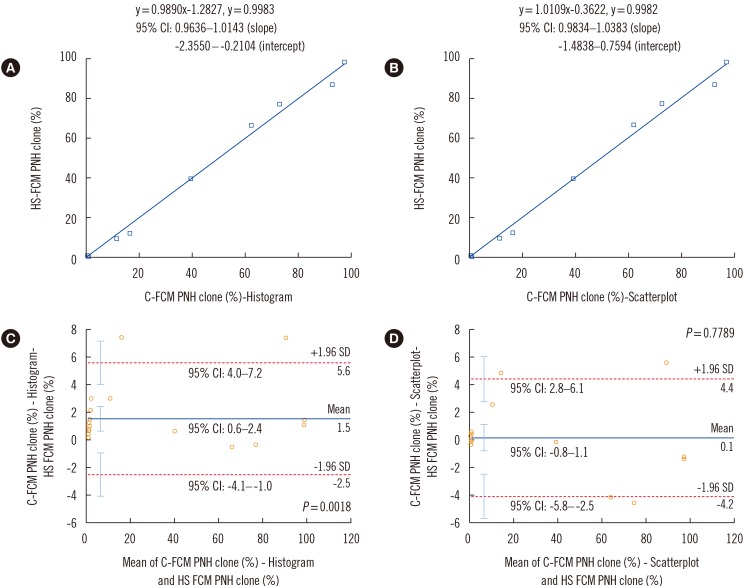 | Fig. 3Comparison of C-FCM RBC PNH clone size results with scatterplot and histogram and HS-FCM. Results were compared by means of Passing-Bablok regression analysis (A-B) and Bland-Altman analysis (C-D). Regression equations, correlation coefficients, and 95% CIs for the slope and intercept of the equations are shown in (A) and (B). In the Bland-Altman analysis, mean difference (indicated by the blue horizontal line), lower and upper limits of agreement (indicated by the red dotted horizontal lines), and their 95% CIs (indicated by the cyan vertical bars) are presented in (C) and (D).Abbreviations: RBC, red blood cell; HS-FCM, high sensitivity flow cytometry; C-FCM, conventional flow cytometry; PNH, paroxysmal nocturnal hemoglobinuria; CI, confidence interval.
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download