1. Meintker L, Ringwald J, Rauh M, Krause SW. Comparison of automated differential blood cell counts from Abbott Sapphire, Siemens Advia 120, Beckman Coulter DxH 800, and Sysmex. XE-2100 in normal and pathological samples. Am J Clin Pathol. 2013; 139:641–650. PMID:
23596116.
2. Eilertsen H, Vøllestad NK, Hagve TA. The usefulness of blast flags on the Sysmex XE-5000 is questionable. Am J Clin Pathol. 2013; 139:633–640. PMID:
23596115.
3. Barnes PW, McFadden SL, Machin SJ, Simson E. International Consensus Group For Hematology. The International Consensus Group for Hematology review: suggested criteria for action following automated CBC and WBC differential analysis. Lab Hematol. 2005; 11:83–90. PMID:
16024331.
4. Novis DA, Walsh M, Wilkinson D, St Louis M, Ben-Ezra J. Laboratory productivity and the rate of manual peripheral blood smear review: a College of American Pathologists Q-Probes study of 95,141 complete blood count determinations performed in 263 institutions. Arch Pathol Lab Med. 2006; 130:596–601. PMID:
16683868.
5. Kim AH, Lee W, Kim M, Kim Y, Han K. White blood cell differential counts in severely leukopenic samples: a comparative analysis of different solutions available in modern laboratory hematology. Blood Res. 2014; 49:120–126. PMID:
25025014.
6. Faucher JL, Lacronique-Gazaille C, Frebet E, Trimoreau F, Donnard M, Bordessoule D, et al. “6 markers/5 colors” extended white blood cell differential by flow cytometry. Cytometry A. 2007; 71:934–944. PMID:
17879238.
7. Jo Y, Kim SH, Koh K, Park J, Shim YB, Lim J, et al. Reliable, accurate determination of the leukocyte differential of leukopenic samples by using Hematoflow method. Korean J Lab Med. 2011; 31:131–137. PMID:
21779183.
8. Kahng J, Kim Y, Kim M, Oh EJ, Park YJ, Han K. Flow cytometric white blood cell differential using CytoDiff is excellent for counting blasts. Ann Lab Med. 2015; 35:28–34. PMID:
25553277.
9. Cossarizza A, De Biasi S, Gibellini L, Bianchini E, Bartolomeo R, Nasi M, et al. Cytometry, immunology, and HIV infection: three decades of strong interactions. Cytometry A. 2013; 83:680–691. PMID:
23788450.
10. Ratei R, Karawajew L, Lacombe F, Jagoda K, Del Poeta G, Kraan J, et al. Discriminant function analysis as decision support system for the diagnosis of acute leukemia with a minimal four color screening panel and multiparameter flow cytometry immunophenotyping. Leukemia. 2007; 21:1204–1211. PMID:
17410192.
11. Allou K, Vial JP, Bene MC, Lacombe F. The routine leukocyte differential flow cytometry HematoFlow method: a new flagging system for automatic validation. Cytometry B Clin Cytom. 2015; 88:375–384. PMID:
25906976.
12. Im M, Chae H, Kim T, Park HH, Lim J, Oh EJ, et al. Comparative quantitative analysis of cluster of differentiation 45 antigen expression on lymphocyte subsets. Korean J Lab Med. 2011; 31:148–153. PMID:
21779186.
13. Buhlmann L, Buser AS, Cantoni N, Gerull S, Tichelli A, Gratwohl A, et al. Lymphocyte subset recovery and outcome after T-cell replete allogeneic hematopoietic SCT. Bone Marrow Transplant. 2011; 46:1357–1362. PMID:
21113185.
14. Fernandez-Ruiz M, Silva JT, Lopez-Medrano F, Allende LM, San Juan R, Cambra F, et al. Post-transplant monitoring of NK cell counts as a simple approach to predict the occurrence of opportunistic infection in liver transplant recipients. Transpl Infect Dis. 2016; 18:552–565. PMID:
27260953.
15. Fujimaki K, Maruta A, Yoshida M, Kodama F, Matsuzaki M, Fujisawa S, et al. Immune reconstitution assessed during five years after allogeneic bone marrow transplantation. Bone Marrow Transplant. 2001; 27:1275–1281. PMID:
11548845.
16. Rueff J, Medinger M, Heim D, Passweg J, Stern M. Lymphocyte subset recovery and outcome after autologous hematopoietic stem cell transplantation for plasma cell myeloma. Biol Blood Marrow Transplant. 2014; 20:896–899. PMID:
24631739.
17. van de Berg PJ, Hoevenaars EC, Yong SL, van Donselaar-van der Pant KA, vanTellingen A, Florquin S, et al. Circulating lymphocyte subsets in different clinical situations after renal transplantation. Immunology. 2012; 136:198–207. PMID:
22321054.
18. Ziegler Heitbrock L. The CD14+ CD16+ blood monocytes: their role in infection and inflammation. J Leukoc Biol. 2007; 81:584–592. PMID:
17135573.
19. Kalina T, Flores-Montero J, Lecrevisse Q, Pedreira CE, van der Velden VH, Novakova M, et al. Quality assessment program for EuroFlow protocols: summary results of four-year (2010-2013) quality assurance rounds. Cytometry A. 2015; 87:145–156. PMID:
25345353.
20. Matarraz S, Almeida J, Flores-Montero J, Lecrevisse Q, Guerri V, Lopez A, et al. Introduction to the diagnosis and classification of monocytic-lineage leukemias by flow cytometry. Cytometry B Clin Cytom. 2017; 92:218–227. PMID:
26282340.
21. van Dongen JJ, Lhermitte L, Bottcher S, Almeida J, van der Velden VH, Flores-Montero J, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012; 26:1908–1975. PMID:
22552007.
22. Zare H, Bashashati A, Kridel R, Aghaeepour N, Haffari G, Connors JM, et al. Automated analysis of multidimensional flow cytometry data improves diagnostic accuracy between mantle cell lymphoma and small lymphocytic lymphoma. Am J Clin Pathol. 2012; 137:75–85. PMID:
22180480.
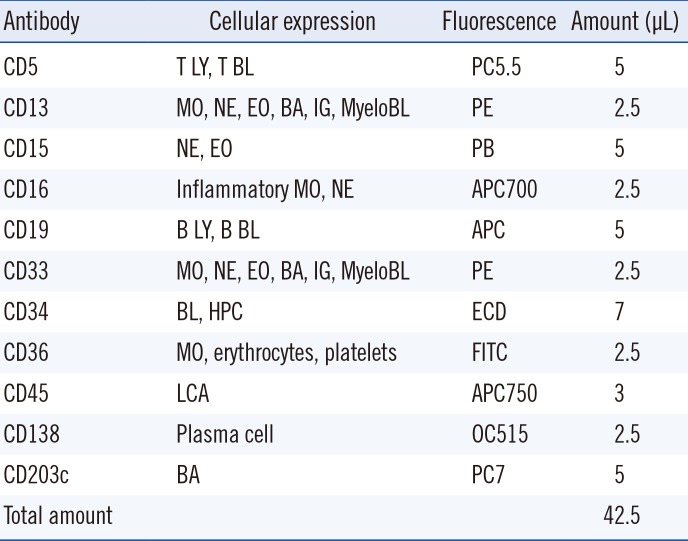
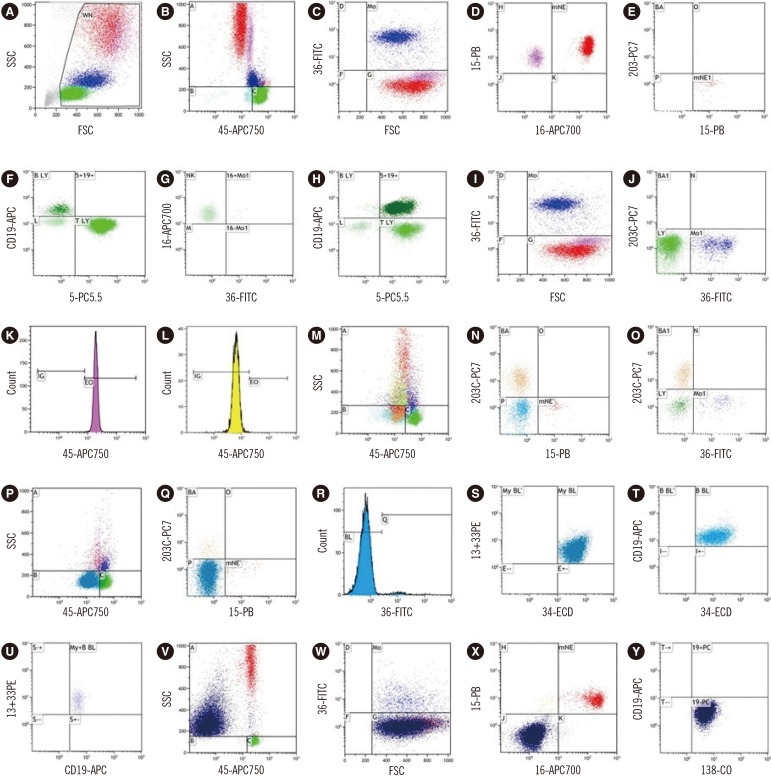
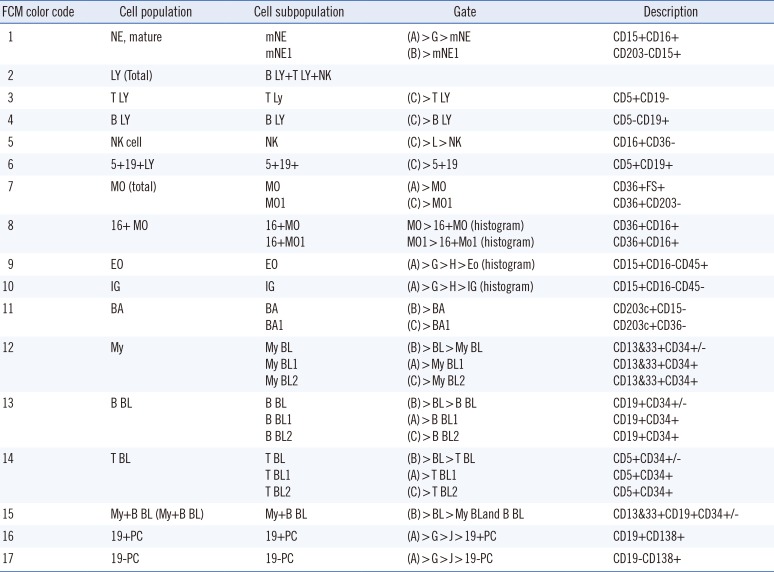
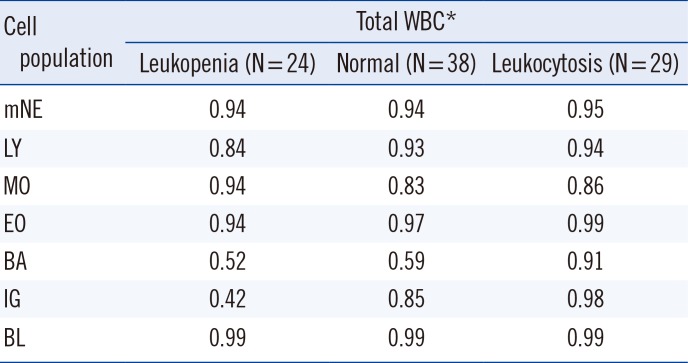




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download