Introduction
Experimental
General experimental procedures
“Black-box” co-culture method
Extraction and isolation of compounds
CD computational methods
X-ray structure determinations
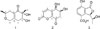
Chinoketide A (1)
 : −35 (c 0.1, CH3OH); UV(CH3OH): λmax (log ε) 245.3 (5.4) nm; Infra-Red (IR) (film): νmax 3215, 2930, 1680, 1450, 1358 cm−1; 1H and 13C NMR data (see Table 1); HRESIMS m/ e 213.1124 [M+H]+ (Calcd. for C11H17O4, 213.1127).
: −35 (c 0.1, CH3OH); UV(CH3OH): λmax (log ε) 245.3 (5.4) nm; Infra-Red (IR) (film): νmax 3215, 2930, 1680, 1450, 1358 cm−1; 1H and 13C NMR data (see Table 1); HRESIMS m/ e 213.1124 [M+H]+ (Calcd. for C11H17O4, 213.1127).Chinoketide B (2)
 = +32 (c 0.1, CH3OH); UV (CH3OH) λmax (log ε) 218.5 (5.4), 231.3 (3.8), 282.7 (5.7) nm; IR (film): νmax 3218, 1682, 1658, 1430 cm−1; 1H and 13C NMR data (see Table 1). HREIMS m/e 196.0374 [M]+ (calcd. for C9H8O5, 196.0372).
= +32 (c 0.1, CH3OH); UV (CH3OH) λmax (log ε) 218.5 (5.4), 231.3 (3.8), 282.7 (5.7) nm; IR (film): νmax 3218, 1682, 1658, 1430 cm−1; 1H and 13C NMR data (see Table 1). HREIMS m/e 196.0374 [M]+ (calcd. for C9H8O5, 196.0372).Xylarphthalide A (3)
 : +65 (c 0.1, CH3OH); m.p. 218 – 219℃. UV (CH3OH): λmax (log ε) 224.6 (5.6), 251.2 (4.4), 302.2 (1.2) nm; IR (film): νmax 3363, 3205, 2925, 1667, 1650, 1609, 1495, 1340 cm−1; 1H NMR (400 MHz in CD3OD-d4): δ 8.15 (d, 8.4, 1H), 6.94 (d, 8.4, 1H), 5.76 (s, 1H), 4.60 (dd, 6.2, 12.7, 1H), 1.45 (d, 6.5,3H); 13C NMR (100 MHz in CD3OD-d4): δ 20.9 (CH3), 67.2 (CH), 86.7 (CH), 114.7 (C), 117.1 (CH), 118.4 (C), 139.6 (CH), 154.0 (C), 161.8 (C), 167.5 (C), 171.5 (C); HRESIMS: m/e 237.0402 [M−H]− (Calcd. for C11H9O6, 237.0399).
: +65 (c 0.1, CH3OH); m.p. 218 – 219℃. UV (CH3OH): λmax (log ε) 224.6 (5.6), 251.2 (4.4), 302.2 (1.2) nm; IR (film): νmax 3363, 3205, 2925, 1667, 1650, 1609, 1495, 1340 cm−1; 1H NMR (400 MHz in CD3OD-d4): δ 8.15 (d, 8.4, 1H), 6.94 (d, 8.4, 1H), 5.76 (s, 1H), 4.60 (dd, 6.2, 12.7, 1H), 1.45 (d, 6.5,3H); 13C NMR (100 MHz in CD3OD-d4): δ 20.9 (CH3), 67.2 (CH), 86.7 (CH), 114.7 (C), 117.1 (CH), 118.4 (C), 139.6 (CH), 154.0 (C), 161.8 (C), 167.5 (C), 171.5 (C); HRESIMS: m/e 237.0402 [M−H]− (Calcd. for C11H9O6, 237.0399).Antimicrobial activity
Result and Discussion
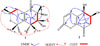
 = −35 (c 0.1, CH3OH). Its UV spectrum displayed maximum absorption peaks at 245.3 nm. Its molecular formula was determined as C11H16O4 by the pseudomolecular ion peak at [M+H]+ m/e 213.1124 (calcd. 213.1127) in the HRESIMS spectrum. The 1H and 13C NMR spectra of 1 showed seven groups of proton signals and eleven carbon resonance signals. In the 13C and DEPT NMR spectra, there were four quaternary carbon signals at δ 199.4, 151.9, 127.4 and 76.4, two methine groups at δ 71.4 and 68.8, three methylene groups at δ 63.1, 36.9 and 36.7, and two methyl groups at δ 21.0 and 18.1. In the 1H -1H COSY spectrum, the cross peak between δ 3.73 (m, 1H) (H-3) and 2.52 (m, 1H)/2.18 (m, 1H) (H-4) proved that H-3 was adjacent to H-4, and the proton signal at δ 3.52 (m, 1H) correlated to two proton signals at 1.16 (d, 6.1, 3H) (H-9) and 2.09 (m, 2H) (H-7), suggesting the existence of substructure of −CH2-CH(OR)-CH3. Following comprehensive analysis of the HSQC and HMBC spectra of compound 1, the planar structure of compound 1 was built up (shown in Fig. 1). In the HMBC spectrum of compound 1, H-10 correlated with C-1, C-2 and C-3, whereas H-3 correlated with C-1, C-2, C-4, C-5 and C-10, on the other hand, H-4 correlated with C-2, C-3, C-5 and C-6 indicating the presence of a six-membered ring with an α,β-unsaturated ketone core in chinoketide A. The cross peaks between H-11 and C-4, C-5, C-6 and between H-4 and C-5, C-6 suggested that C-11 was attached to an α,β-unsaturated ketone core at C-5. And H-7 correlated with C-1, C-5 and C-6, H-8 correlated with C-6 proving that C-7 was connected to C-6 of the six-membered ring. Then, the correlations from H-7 to C-8, C-9, from H-8 to C-7, C-9, from H-9 to C-8, C-9 verified the relative positions of C-7, C-8 and C-9. Finally, the correlations from H-8 to C-11 and from H-11 to C-8 proved that C-8 and C-11 were linked together to form a second six-membered ring in compound 1 via an ether bond. Thus, the structure of chinoketide A was established as shown in Fig. 1.
= −35 (c 0.1, CH3OH). Its UV spectrum displayed maximum absorption peaks at 245.3 nm. Its molecular formula was determined as C11H16O4 by the pseudomolecular ion peak at [M+H]+ m/e 213.1124 (calcd. 213.1127) in the HRESIMS spectrum. The 1H and 13C NMR spectra of 1 showed seven groups of proton signals and eleven carbon resonance signals. In the 13C and DEPT NMR spectra, there were four quaternary carbon signals at δ 199.4, 151.9, 127.4 and 76.4, two methine groups at δ 71.4 and 68.8, three methylene groups at δ 63.1, 36.9 and 36.7, and two methyl groups at δ 21.0 and 18.1. In the 1H -1H COSY spectrum, the cross peak between δ 3.73 (m, 1H) (H-3) and 2.52 (m, 1H)/2.18 (m, 1H) (H-4) proved that H-3 was adjacent to H-4, and the proton signal at δ 3.52 (m, 1H) correlated to two proton signals at 1.16 (d, 6.1, 3H) (H-9) and 2.09 (m, 2H) (H-7), suggesting the existence of substructure of −CH2-CH(OR)-CH3. Following comprehensive analysis of the HSQC and HMBC spectra of compound 1, the planar structure of compound 1 was built up (shown in Fig. 1). In the HMBC spectrum of compound 1, H-10 correlated with C-1, C-2 and C-3, whereas H-3 correlated with C-1, C-2, C-4, C-5 and C-10, on the other hand, H-4 correlated with C-2, C-3, C-5 and C-6 indicating the presence of a six-membered ring with an α,β-unsaturated ketone core in chinoketide A. The cross peaks between H-11 and C-4, C-5, C-6 and between H-4 and C-5, C-6 suggested that C-11 was attached to an α,β-unsaturated ketone core at C-5. And H-7 correlated with C-1, C-5 and C-6, H-8 correlated with C-6 proving that C-7 was connected to C-6 of the six-membered ring. Then, the correlations from H-7 to C-8, C-9, from H-8 to C-7, C-9, from H-9 to C-8, C-9 verified the relative positions of C-7, C-8 and C-9. Finally, the correlations from H-8 to C-11 and from H-11 to C-8 proved that C-8 and C-11 were linked together to form a second six-membered ring in compound 1 via an ether bond. Thus, the structure of chinoketide A was established as shown in Fig. 1. = +32 (c 0.1, CH3OH). Its UV spectrum displayed maximum absorption peaks at 218.5, 231.3, 282.7 nm. Its molecular formula was determined as C9H8O5 by the molecular ion peak at [M]+ m/e 196.0374 [M]+ (calcd. for C9H8O5, 196.0372) in the HR-EI-MS spectrum. The 1H and 13C NMR spectra were very simple, there were only five groups of proton signals in 1H NMR spectrum, and only nine carbon signals in 13C NMR spectrum. There were three quaternary carbon signals at δ 199.8, 164.6, 147.8 and 108.1, four methine groups at δ 108.2, 101.1, 71.6 and 69.8, one methylene group at δ 43.1 (see Table 1).
= +32 (c 0.1, CH3OH). Its UV spectrum displayed maximum absorption peaks at 218.5, 231.3, 282.7 nm. Its molecular formula was determined as C9H8O5 by the molecular ion peak at [M]+ m/e 196.0374 [M]+ (calcd. for C9H8O5, 196.0372) in the HR-EI-MS spectrum. The 1H and 13C NMR spectra were very simple, there were only five groups of proton signals in 1H NMR spectrum, and only nine carbon signals in 13C NMR spectrum. There were three quaternary carbon signals at δ 199.8, 164.6, 147.8 and 108.1, four methine groups at δ 108.2, 101.1, 71.6 and 69.8, one methylene group at δ 43.1 (see Table 1).



 PDF
PDF ePub
ePub Citation
Citation Print
Print





 XML Download
XML Download