INTRODUCTION
MATERIALS AND METHODS
Table
Subjects' characteristics

Quantitative real-time polymerase chain reaction (qRT-PCR)
IHC staining
Western blotting
Enzyme-linked immunosorbent assay (ELISA)
Measurement of immunoglobulin
Flow cytometric analysis
Dispersed polyp cell (DPC) culture and stimulation
Statistical analysis
RESULTS
Hrd1 is differentially expressed in polyp tissues from patients with ECRS and those with non-ECRS
 | Fig. 1Hrd1 mRNA expression in nasal tissues. Hrd1 (A), CD19 (B), CD20 (C), CD138 (D) and BAFF (E) mRNA levels were significantly enhanced in polyp tissues from patients with ECRS compared with values seen in the other subgroups. Results are expressed as medians (interquartile ranges).
BAFF, B-cell activating factor; ECRS, eosinophilic chronic rhinosinusitis with nasal polyp; non-ECRS, non-eosinophilic chronic rhinosinusitis with nasal polyp; CRSsNP, chronic rhinosinusitis without nasal polyps.
|
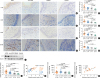 | Fig. 2Increased Hrd1 protein levels and accumulation of B cells and plasma cells in NPs. Representative IHC staining of Hrd1 (A), CD19 (C), CD20 (E), and CD138 (G) (magnification, × 400) in nasal tissues. Relative positive cells of Hrd1 (B), CD19 (D), CD20 (F), and CD138 (H) were significantly increased in polyp tissues from patients with ECRS relative to those in other subgroups. Representative Western blotting results (I), densitometric analysis (J). Positive correlation between relative Hrd1 protein levels in the polyp tissues and Lund-Mackay CT scores of the matched patients (K). ELISA of BAFF in nasal tissues (L). Positive correlation between BAFF and Hrd1 protein levels in the matched NPs (M). Bars = 20 μm.
NP, nasal polyp; ECRS, eosinophilic chronic rhinosinusitis with nasal polyp; non-ECRS, non-eosinophilic chronic rhinosinusitis with nasal polyp; CRSsNP, chronic rhinosinusitis without nasal polyps; HPF, high-power field; CT, computed tomography; ELISA, enzyme-linked immunosorbent assay; BAFF, B-cell activating factor.
|
B-cells and plasma cells are differentially accumulated in NPs from patients with ECRS and those with non-ECRS
Enhanced Hrd1 levels in B-cells in NPs and matched PBMCs
 | Fig. 3Enhanced Hrd1 expression in B-cells from polyp tissues and matched PBMCs. Gating strategy and representative flow plots for nasal tissues (A). MFI levels of Hrd1 in CD19+ B-cells were significantly increased in polyp tissues from patients with ECRS compared with those in in polyps from patients with non-ECRS, middle turbinates from control subjects (B). Gating strategy and representative flow plots for PBMCs (C). MFI levels of Hrd1 in CD19+ B-cells were significantly increased in PBMCs from patients with CRSwNP compared with those in PBMCs from healthy controls (D).
PBMC, peripheral blood mononuclear cell; MFI, mean fluorescence intensity; ECRS, eosinophilic chronic rhinosinusitis with nasal polyp; non-ECRS, non-eosinophilic chronic rhinosinusitis with nasal polyp; CRSwNP, chronic rhinosinusitis with nasal polyps.
|
Increased local immunoglobulin levels in NPs
 | Fig. 4Increased local immunoglobulin levels in polyp tissues. Protein levels of IgA, IgE, IgM, IgG and IgG subclasses in tissue homogenates from ECRS, non-ECRS, CRSsNP, and control tissue are detected by using the Bio-Plex assay. Positive correlations are found between relative Hrd1 protein levels and IgM, IgG, IgG1, and IgG2.
Ig, immunoglobulin; ECRS, eosinophilic chronic rhinosinusitis with nasal polyp; non-ECRS, non-eosinophilic chronic rhinosinusitis with nasal polyp; CRSsNP, chronic rhinosinusitis without nasal polyps.
|
IL-1β, LPS, and Poly(I:C) differentially regulate Hrd1 expression in DPCs from patients with ECRS and those with non-ECRS in vitro
 | Fig. 5Hrd1 mRNA and protein levels in cultured DPCs in response to IL-1β, LPS, Poly(I:C) and Dex. mRNA expression and representative western blot results of DPCs lysates from ECRS patients after 20-hour stimulation (A-C). mRNA expression and representative western blot results of DPCs lysates from non-ECRS patients after 20-hour stimulation (D-F). Results represent mean values from 3 independent experiments. Data are expressed as means (standard error of the means).
DPC, dispersed polyp cell; IL, interleukin; LPS, lipopolysaccharide; Poly(I:C), polyinosinic-polycytidylic acid; Dex, dexamethasone; ECRS, eosinophilic chronic rhinosinusitis with nasal polyp;non-ECRS, non-eosinophilic chronic rhinosinusitis with nasal polyp.
*P < 0.05; †P < 0.01; ‡P < 0.001.
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download