Abstract
We have examined isolation and identification protocols for three virus simulant candidates to biological warfare agents. MS2 phage, a simulant for yellow fever virus and Hantaan virus, was propagated using as a host an E. coli strain with F pilus. MS2 phage genome was examined by reverse transcription and polymerase chain reaction (RT-PCR). Coat protein of the phage preparation was examined by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometric analysis. Cydia pomonella granulosis virus (CpGV) is a virus simulant candidate to smallpox virus. CpGV was isolated from a commercialized CpGV pellet. In this study, we developed new isolation and identification protocols for CpGV. One disadvantage of using CpGV is that it is not easy to determine viability of the virus. Here, we have included T4 phage as an alternative. We established a high titer production protocol and developed an easy genome identification protocol that does not require purified phage DNA. Stability of these virus preparations was also examined under various storage conditions. When the virus preparations were not subjected to freeze drying, MS2 phage was most stable when it was stored in liquid nitrogen but unstable at 4℃. In contrast, T4 phage was most stable when it was stored at 4℃. CpGV was stable at −20℃ but not at 4℃. Stability during or after freeze drying was also investigated. The result showed that 70~80% MS2 survived the freeze drying process. In contrast, only about 15% of T4 phage survived during the freeze drying. CpGV was found to be degraded during freeze drying.
Figures and Tables
Figure 1
Verification of MS2 phage. (A) Reverse transcription (RT)-PCR analysis. RNA was isolated from the purified MS2 sample and used to amplify the phage genome by RT-PCR (lane 2). Lane 1 shows the result of RT-PCR without phage RNA. (B) SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of phage proteins. 18% polyacrylamide gel was used to examine the plate lysate and purified MS2 sample. After electrophoresis, the proteins in the gel were visualized by staining with Coomassie blue. (C) Protein database search after LC-MS/MS. The band indicated by double asterisks in panel B was excised and digested with trypsin. The resulting tryptic peptides were analyzed by LC-MS/MS and protein databases were searched to identify the proteins that matched with the LC-MS/MS result. The panel shown here represents the search result, and the matched peptides are underlined. (D) Electron microscopic examination. Virus sample was mounted on a copper grid and examined under a transmission electron microscope. The virus sample was negatively stained with phosphotungstate.

Figure 2
Purification and identification of CpGV. (A) Purification of CpGV occlusion body by ultracentrifugation. CpGV sample was layered onto discontinuous layers of 60% and 30% sucrose solutions prepared in a swing-out tube and centrifuged at 72,000 g at 4℃ for 1 hr 30 min. Three bands formed after centrifugation were labeled #1, #2, and #3. (B) SDS-PAGE analysis. 12.5% SDS-polyacrylamide gel was used to examine the protein in the band shown in panel A. (C) Protein database search after LC-MS/MS. The major band after Coomassie staining was excised and analyzed as described in Fig. 1C. The result of the protein database search was summarized.
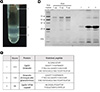
Figure 3
Analysis of CpGV DNA. (A) Agarose gel electrophoresis of purified CpGV DNA. CpGV DNA was electrophoresed in a 0.8% agarose gel and stained with EtBr. (B) Restriction enzyme digestion pattern of CpGV DNA. Virus genomic DNA was digested with either ApaLI or MfeI and the restriction fragments were separated by agarose gel electrophoresis.
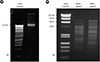
Figure 4
Verification of T4 phage. (A) PCR analysis. DNA was isolated from purified T4 phage sample and used to amplify the phage genome by PCR. (B) Plaque PCR. Purified T4 phage sample was plated with host bacteria and the resulting plaques were randomly taken by poking each plaque with a micropipette tip. The agar plug in the tip was resuspended in distilled water and boiled for 5 min. After centrifugation, 1 µl of the supernatant was directly used in PCR. (C) Electron microscopic examination. Virus sample was examined as described in Fig. 1D.
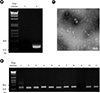
Table 1
Stability of purified MS2 and T4 phages at various storage conditions

a Viability was determined after 5 months. The titer before storage was taken as 100%.
b MS2 phage was stored in liquid nitrogen, and the titer was determined after a given period of months (mo) or years (yr) shown in parenthesis. T4 phage was stored at 4℃ and the titer determination was carried out four times after a given period of time. The titer before storage was taken as 100%.
References
1. Gubser C, Hué S, Kellam P, Smith GL. Poxvirus genomes: a phylogenetic analysis. J Gen Virol. 2004; 85:105–117.

2. Wasserman S, Tambyah PA, Lim PL. Yellow fever cases in Asia: primed for an epidemic. Int J Infect Dis. 2016; 48:98–103.

4. Bentahir M, Laduron F, Irenge L, Ambroise J, Gala JL. Rapid and efficient filtration-based procedure for separation and safe analysis of CBRN mixed samples. PLos One. 2014; 9:e88055.

5. Dawson DJ, Paish A, Staffell LM, Seymour IJ, Appleton H. Survival of viruses on fresh produce, using MS2 as a surrogate for norovirus. J Appl Microbiol. 2005; 98:203–209.

6. Garnier L, Gaudin JC, Bensadoun P, Rebillat I, Morel Y. Real-time PCR assay for detection of a new simulant for poxvirus biothreat agents. Appl Environ Microbiol. 2009; 75:1614–1620.

7. Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogeneis of bacteriophage T4. Cell Mol Life Sci. 2003; 60:2356–2370.
8. Grades ZE. Novel isolates of Cydia pomonella granulo virus (CpGV): deciphering the molecular mechanism for overcoming CpGV resistance in codling moth (Cydia pomonella). Germany: Johannes Gutenberg-Universität Mainz;2010. Ph.D. thesis.
9. Kuzmanovic DA, Elashvili I, Wick C, O'Connell C, Krueger S. Bacteriophage MS2: Molecular weight and spatial distribution of the protein and RNA components by small-angle neutron scattering and virus counting. Structure. 2003; 11:1339–1348.
10. Winstanley D, Crook NE. Replication of Cydia pomonella granulosis virus in cell cultures. J Gen Virol. 1993; 74:1599–1609.
11. Jończyk E, Klak M, Międzybrodzki R, Górski A. The infuence of external factors on bacteriophages-review. Folia Microbiol. 2011; 56:191–200.

12. Ackermann HW, Tremblay D, Moineau S. Long-term bacteriophage preservation. WFCC Newslett. 2004; 38:35–40.
13. Hilton S, Winstanley D. Identification and functional analysis of the origins of DNA replication in the Cydia pomonella granulo virus genome. J Gen Virol. 2007; 88:1496–1504.

14. Gebhardt MM, Eberle KE, Radtke P, Jehle JA. Baculovirus resistance in codling moth is virus isolate-dependent and the consequence of a mutation in viral gene pe38. Proc Natl Acad Sci U S A. 2014; 111:15711–15716.

15. Merabishvili M, Vervaet C, Pirnay JP, Vos DD, Verbeken G, Mast J, et al. Stability of Staphylococcus aureus phage ISP after freezing-drying (lyophilization). PLoS One. 2013; 8:e68797.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download