Abstract
The detection and quantification of hepatitis B virus (HBV) DNA plays an important role in diagnosing and monitoring HBV infection as well as in assessing the therapeutic response. We compared the analytical performance of a random access, fully automated HBV assay—DxN VERIS Molecular Diagnostics System (Beckman Coulter, Brea, CA, USA)—with that of Abbott RealTime HBV assay (Abbott Laboratories, Des Plaines, IL, USA). The between-day precision of the VERIS assay ranged from 0.92% (mean 4.68 log IU/mL) to 4.15% (mean 2.09 log IU/mL) for pooled sera from HBV patients. HBV DNA levels measured by the VERIS HBV assay correlated with the calculated HBV DNA levels (r2=0.9994; P<0.0001). The lower limit of quantification was estimated as 8.76 IU/mL (Probit analysis, 95% confidence interval: 7.32–12.00 IU/mL). Passing-Bablok regression analysis showed good concordance between the VERIS and RealTime assays for 187 chronic HBV samples (y=−0.2397+0.9712x; r=0.981), as well as for 20 drug-resistant HBV genotype C positive samples (y=−0.5415+0.9954x; r=0.961). The VERIS assay demonstrated performance similar to the RealTime assay and is suitable for high-throughput HBV DNA monitoring in large hospital laboratories.
Hepatitis B virus (HBV) infection is one of the most common chronic viral infections [1], and HBV-infected patients carrying different HBV genotypes could have diverse outcomes. Genotype C, which is the most prevalent HBV genotype in Korea, has a higher incidence of natural antiviral-resistant mutation than genotypes B and D and thus could lead to poorer therapeutic response and outcome [2]. The detection and quantification of HBV DNA are essential for diagnosis, establishing the prognosis of HBV infection, and monitoring the virologic response to antiviral therapy [34]. International clinical practice guidelines recommend the use of sensitive nucleic acid amplification technologies for HBV DNA detection [35]. These methods meet the current clinical criteria for detection of a dynamic range of up to 109 IU/mL with linear quantification and a limit of detection (LOD) of 10–20 IU/mL [6789]. However, the available automated HBV DNA assays with nucleic acid amplification technologies are generally designed for batch testing of multiple samples; thus, it is difficult to provide results to clinicians on a priority basis.
The DxN VERIS Molecular Diagnostics System (Beckman Coulter, Brea, CA, USA) is a fully automated system for quantitative molecular assay that combines sample loading, nucleic acid extraction, reaction preparation, real-time PCR assay using TaqMan probes, and result interpretation [10]. The VERIS HBV assay uses a quantitative nucleic acid-based amplification technology and is calibrated to the third WHO International Standard for HBV (National Institute for Biological Standards and Control 10/164) [11]. All analytical processes are integrated within a fully streamlined workflow and random access system, enabling rapid quantification of HBV DNA. The system allows samples to be assayed as soon as they arrive in the laboratory, throughout the day. This reduces the turnaround time and decreases the hands-on time of medical technicians.
We evaluated the analytical performance of the DxN VERIS HBV assay in comparison with the Abbott RealTime HBV assay (Abbott Laboratories, Des Plaines, IL, USA) in Korean HBV patients with genotype C. This study was approved by the Institutional Review Board of The Catholic University of Korea (KC17TISI0041).
Plasma samples were collected for routine HBV DNA quantification in cases of chronic HBV infection at Seoul St. Mary Hospital, Seoul, Korea, between November 2016 and May 2017 using BD PPT EDTA gel tubes (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Samples left over after the assay were aliquoted into 6–10 microtubes and stored immediately at −20℃ until analysis.
To assess precision, pooled plasma samples containing three different levels (2, 3, and 4 log IU/mL) of HBV DNA and a set of four commercial controls consisting of one negative and three different levels of HBV DNA as positive controls (Amplichek I, Bio-Rad Laboratories, Inc., Hercules, CA, USA) were assayed in duplicate, twice a day for five consecutive days. MedCalc version 17.6 (MedCalc, Ostend, Belgium) was used for statistical analyses, and P<0.05 was considered statistically significant.
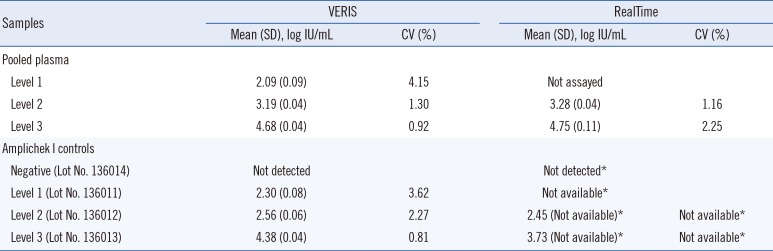
The intra-assay CV of the VERIS assay ranged from 0.46% to 5.66% and 0.36% to 6.33% for pooled plasma and the Amplichek I HBV DNA positive controls, respectively. The inter-assay between-day precision CV ranged from 0.92% to 4.15% and 0.81% to 3.62% for pooled plasma and Amplichek I controls, respectively, suggesting good and similar precision at clinically meaningful levels of <100 IU/mL at the end of anti-viral therapy and >2,000 IU/mL at the time of diagnosis (Table 1).
To evaluate linearity, seven serially diluted pooled plasma samples ranging from 7.18 to 1.18 log IU/mL were measured in triplicate. The levels of difference varied between 0.28 and 0.43 log IU/mL (Fig. 1A).
The lower limit of quantification (LLoQ) was estimated as 8.76 IU/mL (95% confidence interval [CI]: 7.32–12 IU/mL) by Probit analysis using 10 replicates of five levels of pooled plasma samples (approximately 40, 20, 10, 5, and 2.5 IU/mL) in the same experiment. This was equivalent to an LOD of 10 IU/mL (1.00 log IU/mL), according to the manufacturer. Specificity was assessed with eight samples collected from patients with other viral infections (cytomegalovirus, Epstein-Barr virus, hepatitis C virus, and HIV); these were all determined to be negative using the VERIS HBV assay.
For comparative evaluation between the VERIS and RealTime assays, 187 samples ranging from 1.04 to 9.51 log IU/mL (determined using the RealTime assay) were sequentially chosen at random and stored at −20℃ for up to three days before re-analysis. Of these, 57.2% (107/187) and 36.4% (68/187) showed delta differences of <0.5 log IU/mL and 0.5–1.0 log IU/mL, respectively. This result was superior to that of Braun et al. [11] who found that concordance between the two assays was 76% (47.8% and 36.7% of their samples showed delta differences of <0.5 log IU/mL and 0.5–1.0 log IU/mL, respectively). Passing-Bablok regression analysis did not reveal any significant deviation from linearity between the VERIS and RealTime assays (r=0.981; P<0.0001 by Spearman's rank correlation coefficient; linear regression equation, y=−0.2397+0.9712x; P=0.64 by the cumulative sum control chart [CUSUM] test for linearity; Fig. 1B). The mean difference in HBV DNA levels between these two assays was −0.3674 log IU/mL (95% CI, −0.4373 to −0.2974) in the Bland-Altman plot (Fig. 1C). Braun et al. [11] also demonstrated a strong correlation between the VERIS and RealTime assays (r=0.933).
To assess the influence of HBV drug resistance mutations on HBV DNA quantification, plasma samples from 20 patients with drug-resistant HBV genotype C infection were assayed in parallel using the VERIS and RealTime assays. Both assays showed a strong correlation (r=0.961; P<0.0001; linear regression equation, y=−0.5415+0.9954x; P=0.36) for the 20 drug-resistant HBV DNA samples, with a mean difference of −0.4425 log IU/mL (95% CI, −1.3985 to 0.5135). We confirmed good concordance between the VERIS and RealTime assays in Korean patients with HBV genotype C. In contrast, in Braun et al.'s study [11], the readings obtained from the Roche COBAS TaqMan HBV test (Roche Molecular Systems, Pleasanton, CA, USA) and VERIS were higher than expected for genotype D and lower than expected for genotype A (to a lesser extent), with genotypes D and A being the most prevalent genotypes in Europe.
Although several factors, including quality and stability of reference standards, quality and stability of reagents, and statistical validity of the calibration curve may lead to assay discrepancies, the negative bias was consistent across the dynamic range of DNA quantification for the VERIS assay compared with the RealTime assay, which is similar to the results (−0.39 log IU/mL) of Braun et al. [11]. This finding could influence treatment decisions at HBV DNA levels of <2,000 IU/mL according to the Korean Association for the Study of the Liver guidelines for immune-active phase chronic hepatitis B [12] because of the underestimation of DNA quantification. However, treatment decisions are based on various factors, such as patient age, liver enzyme levels, liver histopathology, previous treatment history, and family history [5]. This consideration of multiple factors lessens the impact of the negative bias observed with the VERIS HBV assay.
We showed that the VERIS assay has good precision and excellent analytic sensitivity, with an estimated LLoQ equivalent to that claimed by the manufacturer. Furthermore, we demonstrated that it accurately quantifies HBV DNA levels in clinical samples. Its performance appears to be similar to that of the RealTime assay, a real-time PCR platform widely used in clinical practice in HBV genotype C. Although the VERIS assay will not be available in the future in the automated assay market, many laboratories worldwide, including those in Korea, still use this assay for clinical samples. Our performance data can also serve as a future benchmark for random access, automated viral nucleic acid detection assays, as well as for HBV DNA assays [4131415].
One limitation of our study is that the influence of the different HBV genotypes on HBV DNA quantification was not assessed. We assayed only 20 drug-resistant HBV genotype C positive samples and confirmed the previous results by Fourati et al. [10], who demonstrated a median difference of −0.32 log IU/mL between the VERIS HBV and Cobas AmpliPrep/Cobas TaqMan 96 HBV v2.0 assays for genotype C. The prevailing HBV genotypes in Korea are genotype C2 or a mixed pattern of genotypes B and C, while other genotypes rarely occur [16]. Therefore, our performance data are not applicable for detecting HBV DNA with different genotypes. In conclusion, the VERIS assay demonstrated performance similar to the RealTime assay and is suitable for high-throughput HBV DNA monitoring in large hospital laboratories.
Acknowledgment
The VERIS hepatitis B virus (HBV) kits used in this study were provided by Beckman Coulter (Seoul, Korea); however, the company was not involved in the study design or manuscript preparation. This work was supported by the Technology Innovation Program (No: 10049771, Development of Highly-Specialized Platform for IVD Medical Devices) funded by the Ministry of Trade, Industry & Energy, Korea.
References
1. Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015; 386:1546–1555. PMID: 26231459.
2. Zhang Q, Liao Y, Cai B, Li Y, Li L, Zhang J, et al. Incidence of natural resistance mutations in naïve chronic hepatitis B patients: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2015; 30:252–261. PMID: 25318660.
3. European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017; 67:370–398. PMID: 28427875.
4. Han MS, Park Y, Nah H, Kim HS. Comparison of the QIAGEN artus HBV QS-RGQ assay with the Roche COBAS AmpliPrep/COBAS TaqMan HBV assay for quantifying viral DNA in sera of chronic Hepatitis B patients. Ann Lab Med. 2017; 37:248–253. PMID: 28224771.
5. Terrault NA, Bzowej NH, Chang KM, Hwang JP, Jonas MM, Murad MH, et al. AASLD guidelines for treatment of chronic hepatitis B. Hepatology. 2016; 63:261–283. PMID: 26566064.
6. Pawlotsky JM, Dusheiko G, Hatzakis A, Lau D, Lau G, Liang TJ, et al. Virologic monitoring of hepatitis B virus therapy in clinical trials and practice: recommendations for a standardized approach. Gastroenterology. 2008; 134:405–415. PMID: 18242209.
7. Chevaliez S, Bouvier-Alias M, Laperche S, Hézode C, Pawlotsky JM. Performance of version 2.0 of the Cobas AmpliPrep/Cobas TaqMan Real-time PCR assay for hepatitis B virus DNA quantification. J Clin Microbiol. 2010; 48:3641–3647. PMID: 20720031.
8. Chevaliez S, Rodriguez C, Pawlotsky JM. New virologic tools for management of chronic hepatitis B and C. Gastroenterology. 2012; 142:1303–1313.e1. PMID: 22537437.
9. Yeh ML, Huang CF, Hsieh MY, Huang JF, Dai CY, Yu ML, et al. Comparison of the Abbott RealTime HBV assay with the Roche Cobas AmpliPrep/Cobas TaqMan HBV assay for HBV DNA detection and quantification. J Clin Virol. 2014; 60:206–214. PMID: 24809730.
10. Fourati S, Challine D, Poveda JD, Laperche S, Rallier S, Pawlotsky JM, et al. Evaluation of a new random-access HBV DNA molecular assay: The VERIS HBV assay. J Clin Virol. 2017; 92:69–74. PMID: 28549336.
11. Braun P, Delgado R, Drago M, Fanti D, Fleury H, Izopet J, et al. A European multicentre study on the comparison of HBV viral loads between VERIS HBV assay and Roche COBAS® TAQMAN® HBV test, Abbott RealTime HBV assay, Siemens VERSANT HBV assay, and Qiagen artus HBV RG kit. J Clin Virol. 2017; 95:76–83. PMID: 28892764.
12. The Korean Association for the Study of the Liver (KASL). KASL clinical practice guidelines: management of chronic hepatitis B. Clin Mol Hepatol. 2016; 22:18–75. PMID: 27044762.
13. Huh HJ, Park JE, Kim JY, Yun SA, Lee MK, Lee NY, et al. Performance of the Real-Q EBV quantification kit for Epstein-Barr virus DNA quantification in whole blood. Ann Lab Med. 2017; 37:147–150. PMID: 28029001.
14. Kim H, Hur M, Kim JY, Moon HW, Yun YM, Cho HC. Automated nucleic acid extraction systems for detecting cytomegalovirus and Epstein-Barr virus using real-time PCR: a comparison study between the QIAsymphony RGQ and QIAcube systems. Ann Lab Med. 2017; 37:129–136. PMID: 28028999.
15. Park JE, Kim JY, Yun SA, Lee MK, Huh HJ, Kim JW, et al. Performance evaluation of the Real-Q cytomegalovirus (CMV) quantification kit using two real-time PCR systems for quantifying CMV DNA in whole blood. Ann Lab Med. 2016; 36:603–606. PMID: 27578516.
16. Cho JH, Yoon KH, Lee KE, Park DS, Lee YJ, Moon HB, et al. Distribution of hepatitis B virus genotypes in Korea. Korean J Hepatol. 2009; 15:140–147. PMID: 19581766.
Fig. 1
Performance evaluation of the Beckman Coulter DxN VERIS HBV assay in comparison with the Abbott RealTime HBV assay in 187 HBV samples. (A) Linearity analysis of HBV DNA levels using serially diluted samples (1.18–7.18 log IU/mL). HBV DNA levels obtained by the VERIS HBV assay were related to calculated HBV DNA levels. The dashed line represents the equality line. (B) Passing-Bablok regression analysis of the VERIS and RealTime HBV assays. The dashed line represents the equality line, and the dotted lines represent 95% confidence interval. (C) Bland-Altman plot analysis of the VERIS and RealTime HBV assays. The straight and dashed lines both represent mean differences±1.96 SD.
Abbreviation: HBV, hepatitis B virus.
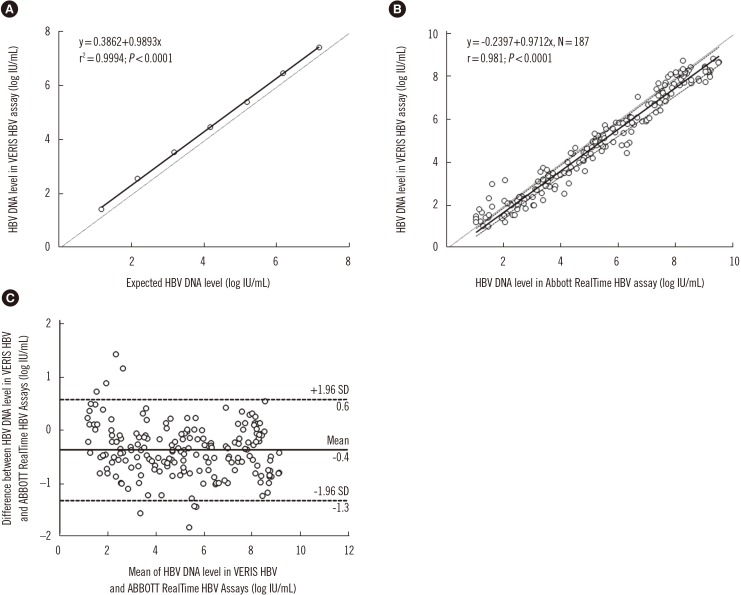
Table 1
Results of between-day precision of the VERIS and RealTime hepatitis B virus assays




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download