Abstract
Background
Methods
Results
Acknowledgment
Notes
Authors' Disclosures of Potential Conflicts of Interest: The authors declare that they have no conflict of interest.
Author Contributions: Zhan-qing Zhang conceived and designed the study. Yan-bing Wang and Wei Lu collated the data. Dan-ping Liu, Bi-sheng Shi, and Xiao-nan Zhang conducted the experiments. Zhan-qing Zhang analyzed the data. Yan-bing Wang, Wei Lu, Dan Huang, Xiu-fen Li, Xin-lan Zhou, Rong-rong Ding, and Zhan-qing Zhang coordinated the collection of human materials. Zhan-qing Zhang and Dan-ping Liu wrote the manuscript. Zhan-qing Zhang critically revised the manuscript.
References
Fig. 1
Distribution of serum virological markers in the four phases of chronic HBV infection. (A) HBcrAg. (B) HBsAg. (C) HBeAg. (D) HBV DNA. The middle horizontal red line represents the median; the upper and lower horizontal red lines represent the quartiles.
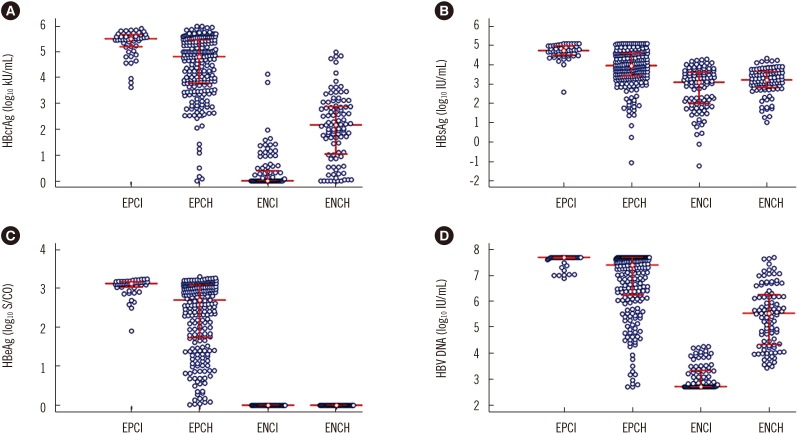
Fig. 2
ROC curves and AUCs of serum virological markers for predicting EPCH and ENCH. (A) ROC curves for predicting EPCH. (B) ROC curves for predicting ENCH. (C) AUCs for predicting EPCH. (D) AUCs for predicting ENCH.
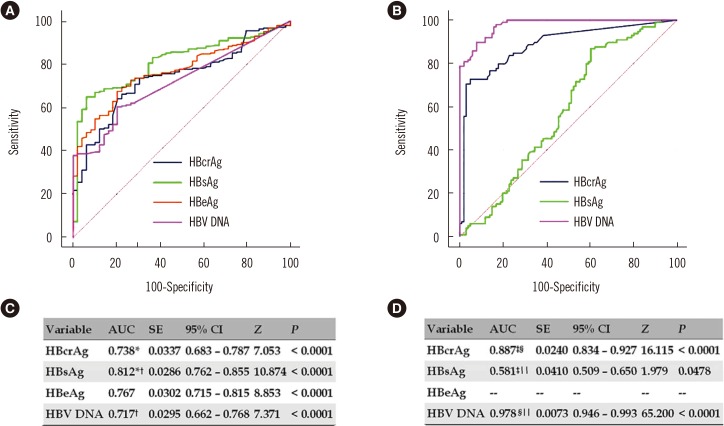
Table 1
Clinical, biochemical, virological, and pathological characteristics of the study population
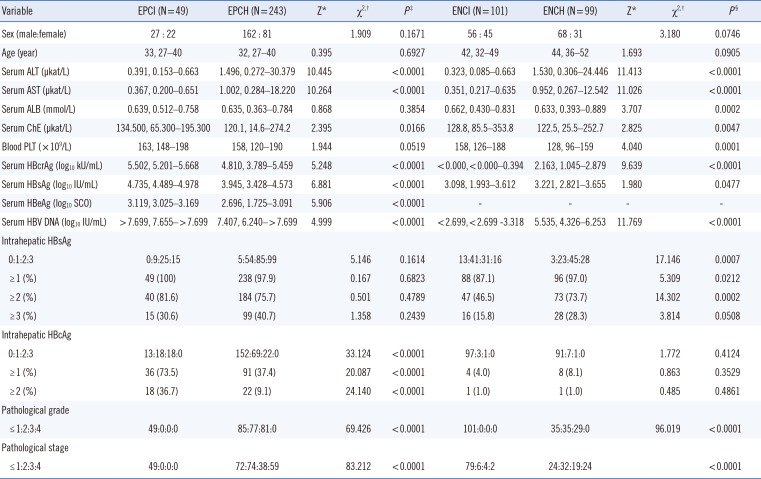
Continous data are presented as median (range),and categorical data are presented as N (%).
*Two-independent samples Mann-Whitney U test; †Pearson's chi-square test; ‡EPCI vs EPCH; §ENCI vs ENCH.
Abbreviations: HBeAg, hepatitis B e antigen; EPCI, HBeAg-positive chronic infection; EPCH, HBeAg-positive chronic hepatitis; ENCI, HBeAg-negative chronic infection; ENCH, HBeAg-negative chronic hepatitis; ALT, alanine transferase; AST, aspartate transferase; ALB, albumin; ChE, cholinesterase; PLT, platelet; HBcrAg, hepatitis B core-related antigen; HBsAg, hepatitis B surface antigen; HBV DNA, hepatitis B virus DNA; HBcrAg, hepatitis B core antigen.
Table 2
Spearman's correlation coefficients of HBcrAg with biochemical parameters, other virological markers, and pathological states
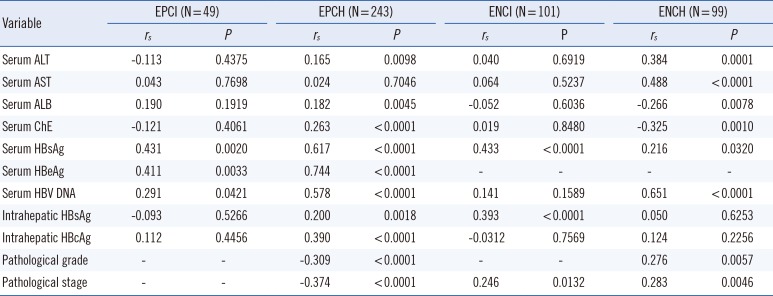
Abbreviations: HBeAg, hepatitis B e antigen; EPCI, HBeAg-positive chronic infection; EPCH, HBeAg-positive chronic hepatitis; ENCI, HBeAg-negative chronic infection; ENCH, HBeAg-negative chronic hepatitis; ALT, alanine transferase; AST, aspartate transferase; ALB, albumin; ChE, cholinesterase; HBcrAg, hepatitis B core-related antigen; HBsAg, hepatitis B surface antigen; HBV DNA, hepatitis B virus DNA; HBcrAg, hepatitis B core antigen.
Table 3
Reference cutoffs and the corresponding diagnostic parameters of serum virological markers in predicting EPCH and ENCH
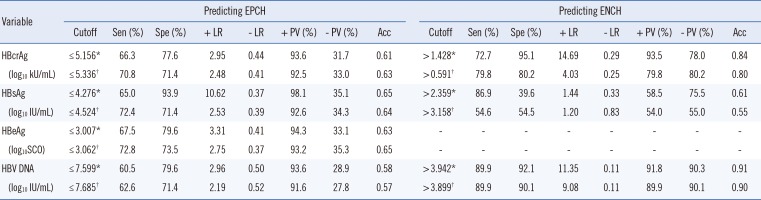
*Optimal cutoffs; †Tradeoff cutoffs.
Abbreviations: HBcrAg, hepatitis B core-related antigen; HBsAg, hepatitis B surface antigen; HBeAg, hepatitis B e antigen; HBV DNA, hepatitis B virus DNA; EPCH, HBeAg-positive chronic hepatitis; ENCH, HBeAg-negative chronic hepatitis; Sen, sensitivity; Spe, specificity; + LR, positive likelihood rate; LR, negative likelihood rate; + PV, positive predictive value; − PV, negative predictive value; acc, accuracy; SCO, signal to cutoff ratio.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download