This article has been
cited by other articles in ScienceCentral.
Abstract
Background
The transition from manual processing of patient samples to automated workflows in medical microbiology is challenging. Although automation enables microbiologists to evaluate all samples following the same incubation period, the essential incubation times have yet to be determined. We defined essential incubation times for detecting methicillin-resistant Staphylococcus aureus (MRSA), multi-drug resistant gram-negative bacteria (MDRGN), and vancomycin-resistant enterococci (VRE).
Methods
We monitored the growth kinetics of MRSA, MDRGN, and VRE between two and 48 hours on chromogenic media to establish the time points of first growth, single colony appearance, and typical morphology for 102, 104, 106, and 108 colony forming units/mL. Subsequently, we imaged plates inoculated with 778 patient samples after 20, 24, and 36 hours.
Results
The first growth, single colony appearance, and typical morphology time points were inoculum-dependent. First growth appeared after 6–18 hours, 4–18 hours, and 8–48 hours for MRSA, MDRGN, and VRE, respectively, and single colonies appeared at 12–18 hours, 6–20 hours, and 12–48 hours, respectively. Typical morphology was visible at 14–22 hours and 12–48 hours for MRSA and VRE, but was not determined for MDRGN. By examining patient samples, ≥98% of MRSA and MDRGN were visible 20 hours after the start of incubation. Following 24 hours of incubation, only 79.5% of VRE were clearly visible on the respective plates.
Conclusions
An incubation time of 20 hours is sufficient for detecting MRSA and MDRGN. VRE growth is much slower and requires additional imaging after 36 hours.
Go to :

Keywords: Laboratory automation, Methicillin-resistant Staphylococcus aureus, Multi-drug resistant gram-negative bacteria, Vancomycin-resistant enterococci, Chromogenic media, Workflow
INTRODUCTION
Fast and reliable identification of bacterial species and susceptibility testing are important steps in the treatment of patients. This is especially true for the treatment of infectious diseases, as well as for screening for multi drug-resistant bacteria, that is, methicillin-resistant Staphylococcus aureus (MRSA), multi-drug resistant gram-negative bacteria (MDRGN), and vancomycin-resistant enterococci (VRE). In our hospital, patients with a history of living in a nursing home, those with a previous hospital stay, and those from high-incidence countries undergo screening shortly after admission. On the basis of this assessment, they are either preemptively isolated until it is determined that they do not carry MRSA, MDRGN, or VRE, or they are isolated (barrier isolation/single room) once they test positive for MRSA, MDRGN, or VRE. In both cases, rapid diagnosis is crucial to minimize the risk of transmission and to avoid unnecessary isolation, costs, and hands-on time.
In the classic manual workflow, samples being screened were incubated for two days, and two reads were necessary. The samples were sent to the laboratory and inoculated on day 0. On day 1, the plates were read for the first time, and on day 2, the second and final read was conducted to reliably fulfill the incubation times specified in the package inserts. Exact inoculation times are not recorded in the classic manual workflow.
With total laboratory automation (TLA), incubation times can be determined precisely to the hour and minute. However, the recommended incubation times mentioned in the package inserts were determined in a pre-TLA era, when incubation times were defined in days or hours instead of hours and minutes. In our case, these recommended incubation times were 20–26 hours for the chromogenic MRSA plate (MRSA II, BD, Heidelberg, Germany), 18–24 hours for the chromogenic extended spectrum beta-lactamase (ESBL) plate (CHROMID ESBL Agar, bioMérieux SA, Marcy l'Etoile, France), and 24–48 hours for the chromogenic VRE plates (CHROMagar VRE Mast, Diagnostica GmbH, Reinfeld, Germany [24 hours]; VRESelect, BioRad Laboratories GmbH, München, Germany [24–28 hours]; and CHROMID VRE Agar, bioMérieux SA [24–48 hours]). We aimed to determine the incubation times needed for the reliable detection of MRSA, MDRGN, and VRE using the above-mentioned plates in order to establish a workflow with a single reading time point as early as possible, within the first 24 hours of incubation.
Go to :

METHODS
This combined retrospective and prospective study was exempt from Institutional Review Board (IRB) approval because studies for quality control for laboratory assays are generally exempt from IRB approval.
Growth kinetics
The strains used for growth kinetics were confirmed as MRSA, MDRGN, or VRE by the following methods: nuc and mecA PCR for MRSA, susceptibility testing using VITEK 2 AST cards (AST-N248, bioMérieux SA) for MDRGN, and vanA and vanB PCR for VRE.
Four different bacterial concentrations of these strains were used to inoculate the plates; this was done to mimic the different amounts of bacteria in patient samples. First, a 0.5 McFarland solution was prepared and considered to contain 10
8 colony forming units (CFU)/mL bacteria. Three serial 1:100 dilutions were prepared and designated as 10
6, 10
4, and 10
2 CFU/mL. The following plates were used for monitoring growth kinetics: MRSA II, CHROMID ESBL, CHROMagar VRE MAST (6 mg/L vancomycin), VRESelect (8 mg/L vancomycin), and CHROMID VRE (8 mg/L vancomycin). The following plates were used for monitoring patient samples: MRSA II (BD), CHROMID ESBL, and VRESelect (8 mg/L vancomycin). The strains used for monitoring growth kinetics are described in
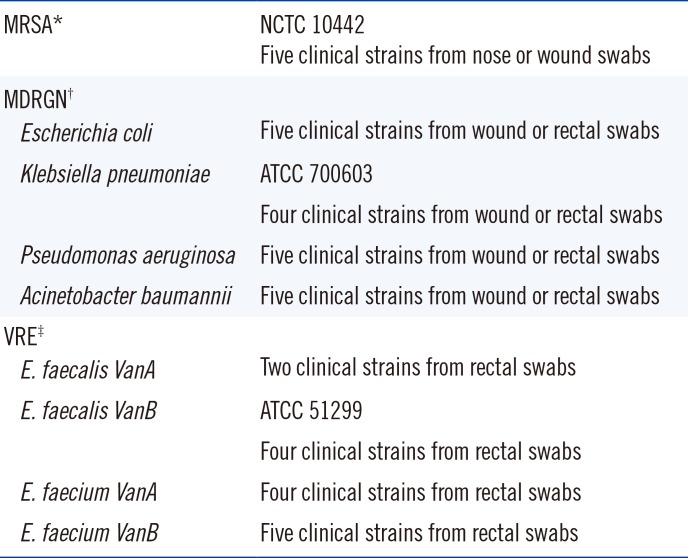
Table 1.
Table 1
Strains used for growth kinetics
|
MRSA*
|
NCTC 10442 |
|
Five clinical strains from nose or wound swabs |
|
MDRGN†
|
|
|
Escherichia coli
|
Five clinical strains from wound or rectal swabs |
|
Klebsiella pneumoniae
|
ATCC 700603 |
|
Four clinical strains from wound or rectal swabs |
|
Pseudomonas aeruginosa
|
Five clinical strains from wound or rectal swabs |
|
Acinetobacter baumannii
|
Five clinical strains from wound or rectal swabs |
|
VRE‡
|
|
|
E. faecalis VanA
|
Two clinical strains from rectal swabs |
|
E. faecalis VanB
|
ATCC 51299 |
|
Four clinical strains from rectal swabs |
|
E. faecium VanA
|
Four clinical strains from rectal swabs |
|
E. faecium VanB
|
Five clinical strains from rectal swabs |

Plates were inoculated with 10 µL of the bacterial solutions using the BD Kiestra TLA system (BD Kiestra, Drachten, Netherlands) and incubated at 35℃ without CO2. MRSA plates were imaged every two hours until 26 hours and then at 36 hours and 48 hours. ESBL plates were imaged every two hours until 24 hours and then at 36 hours and 48 hours. VRE plates were imaged every two hours until 28 hours and then at 36 hours and 48 hours. All imaging was done using the BD Kiestra TLA system. Three time points were determined during these readings: A) the first growth time point, defined as the time point at which growth was first visible; B) the single colonies time point, defined as the time point at which single colonies were discernible and follow-up work was possible (Vitek2 for susceptibility testing and/or matrix assisted laser desorption-time of flight mass spectrometry [MALDI -TOF MS] for identification); and C) the typical morphology time point, defined as the time point when growth and colony morphology were as described in the package inserts (MRSA and VRE plates only).
Evaluation with patient samples
Patient samples used to evaluate the imaging time points consisted of nasal swabs for MRSA and rectal and wound swabs for MRSA, MDRGN, and VRE. All swabs were collected using ESwabs (Copan, Brescia, Italy). Plates were inoculated with 10 µL of patient sample and incubated at 35℃ without CO2. Patient sample evaluation was conducted in two steps. In the first trial, growth after 24 hours and 36 hours was compared. In the second trial, growth after 20 hours and 24 hours was compared with different patient samples. To determine the detection rate at different imaging time points, 95 MRSA-positive samples were reevaluated for growth at 36 hours and 24 hours. Additional 100 MRSA-positive samples were reevaluated for growth at 24 hours and 20 hours. For MDRGN, 153 samples were reevaluated for growth at 36 hours and 24 hours, and additional 352 samples were reevaluated for growth at 24 hours and 20 hours. For VRE, 78 positive samples were evaluated for growth at 36 hours and 24 hours. VRE growth after 20 hours of incubation was not evaluated. A positivity rate of ≥98% at the earlier time point was considered sufficient for reducing routine incubation time. The decision to review the above sample numbers and to accept a maximum loss of 2% of positive samples was an arbitrary decision based on the reading experience of the technicians and on our experience with medical validation of reports over the years.
Go to :

RESULTS
First growth, single colony appearance, and typical morphology on all chromogenic media were dependent on inoculum concentration: the higher the inoculum concentration, the earlier the growth was detected. Certain 102 CFU/mL inocula (one MRSA, eight VRE [VRESelect, BioRad], and four MDRGN) did not result in visible growth.
First growth of MRSA was detected after 6–10 hours if 10
8 CFU/mL inocula were used (
Fig. 1). For the same strains, first growth was visible after 14–18 hours if 10
2 CFU/mL inocula were used (data not shown). Single colonies were detected after 12–14 hours (10
8/mL) and 16–18 hours (10
2/mL). Typical morphology was detected at 14–16 hours (10
8/mL) and 18–22 hours (10
2/mL).
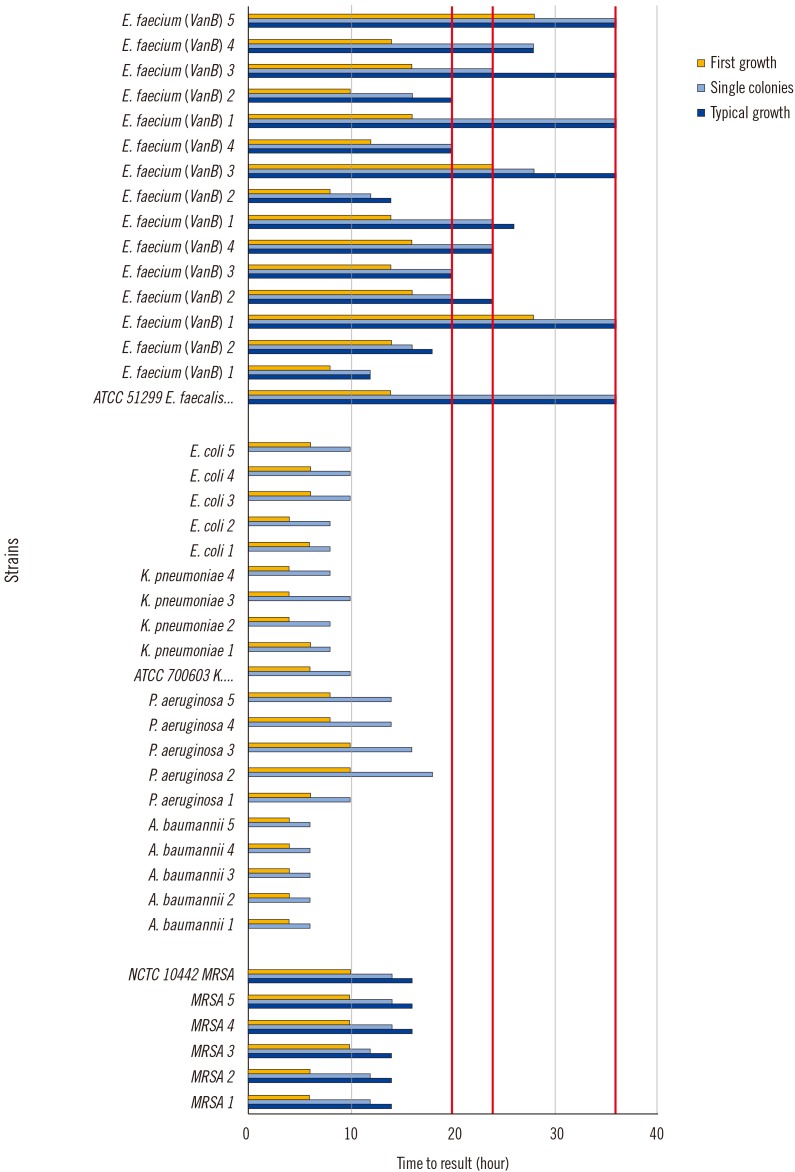 | Fig. 1
Growth kinetics: time to result (=first growth, single colonies, typical morphology) for clinical strains and ATCC strains (MRSA, MDRGN, VRE, 108/mL). Red lines represent imaging times for patient sample evaluation.
Abbreviations: ATCC, American Type Culture Collection; MRSA, methicillin-resistant Staphylococcus aureus; MDRGN, multi-drug resistant gram-negative bacteria; VRE, vancomycin-resistant enterococci.

|
For MDRGN, first growth was detected at 4–10 hours (108/mL) and 10–18 hours (102/mL). Single colonies were detected at 6–18 hours (108/mL) and 10–18 hours (102/mL). Pseudomonas aeruginosa strains tended to appear after Enterobacteriaceae and Acinetobacter baumannii strains. We did not evaluate the plates for typical morphology of gram-negative rods because except for Escherichia coli, all isolates were identified using MALDI-TOF MS or Vitek2 ID panels.
For VRE, we tested plates from three different manufacturers. On VRESelect, first growth was detected at 8–28 hours (10
8/mL) and 16–48 hours (10
2/mL). Single colonies were detected at 12–36 hours (10
8/mL) and 16–48 hours (10
2/mL). Typical morphology was detected at 12–36 hours (10
8/mL) and 18–48 hours (10
2/mL) (
Fig. 1). The first growth, single colonies, and typical morphology data for CHROMagar VRE and CHROMID VRE were similar to those with VRESelect (data not shown).
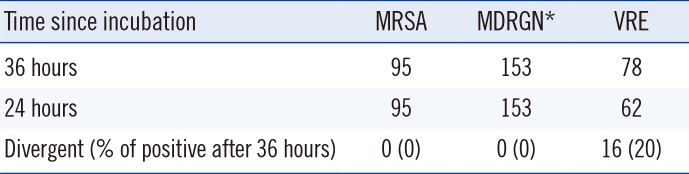
To evaluate patient samples, we commenced imaging after 24 hours and 36 hours (
Table 2). In each case we identified whether growth was already visible at 24 hours or at 36 hours of incubation. In the case of MRSA, all 95 samples showed growth after 24 hours of incubation. For MDRGN, all 153 positive samples evaluated showed clear growth after 24 hours of incubation. For VRE, 62 of 78 (79.5%) were already positive after 24 hours, nine of 78 (11.5%) were only positive after 36 hours of incubation, and in seven of 78 (9%) samples, the colonies were so small after 24 hours of incubation that they could have been easily overlooked. Based on the MRSA and MDRGN results, we changed the imaging times from 24 hours and 36 hours to 20 hours and 24 hours and reevaluated positive samples for first appearance of growth. For MRSA and MDRGN, we evaluated 100 and 352 positive samples, respectively. Of these, 99 MRSA and 351 MDRGN samples showed clearly visible growth after 20 hours of incubation (
Table 3). The sole MRSA sample that showed growth only after 24 hours was a nasal swab, and three colonies were visible after 24 hours. One MDRGN (
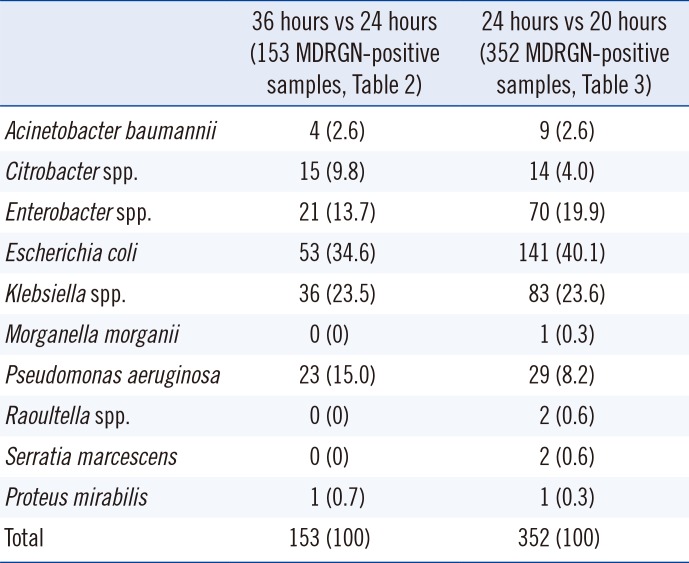
Morganella morganii) sample (rectal swab) exhibited three barely visible white colonies after 20 hours of incubation. The MDRGN species and numbers reevaluated are detailed in
Table 4.
Table 2
Growth in patient samples after 36 hours vs 24 hours of incubation
|
Time since incubation |
MRSA |
MDRGN*
|
VRE |
|
36 hours |
95 |
153 |
78 |
|
24 hours |
95 |
153 |
62 |
|
Divergent (% of positive after 36 hours) |
0 (0) |
0 (0) |
16 (20) |

Table 3
Growth in patient samples after 24 hours vs 20 hours of incubation

|
Time since incubation |
MRSA |
MDRGN*
|
VRE†
|
|
24 hours |
100 |
352 |
- |
|
20 hours |
99 |
351 |
- |
|
Divergent (% of positive after 24 hours) |
1 (1) |
1 (0.3) |
- |

Table 4
Number per species of MDRGN-positive patient samples from Tables 2 and 3
|
36 hours vs 24 hours (153 MDRGN-positive samples, Table 2) |
24 hours vs 20 hours (352 MDRGN-positive samples, Table 3) |
|
Acinetobacter baumannii
|
4 (2.6) |
9 (2.6) |
|
Citrobacter spp. |
15 (9.8) |
14 (4.0) |
|
Enterobacter spp. |
21 (13.7) |
70 (19.9) |
|
Escherichia coli
|
53 (34.6) |
141 (40.1) |
|
Klebsiella spp. |
36 (23.5) |
83 (23.6) |
|
Morganella morganii
|
0 (0) |
1 (0.3) |
|
Pseudomonas aeruginosa
|
23 (15.0) |
29 (8.2) |
|
Raoultella spp. |
0 (0) |
2 (0.6) |
|
Serratia marcescens
|
0 (0) |
2 (0.6) |
|
Proteus mirabilis
|
1 (0.7) |
1 (0.3) |
|
Total |
153 (100) |
352 (100) |

Go to :

DISCUSSION
From a clinician's perspective, laboratory reports tend to take too long; the benefits of rapid reporting of multi-resistant organisms are undisputed. As early as 2001, Rubinovitch and Pittet [
1] stated that successful programs for reducing hospital transmission of MRSA are based on early identification of the MRSA reservoir and prompt implementation of contact precautions. Various PCR techniques have been developed to accelerate reporting [
23], with the drawbacks of high cost, the need for highly qualified personnel, and limited detection of a very narrow spectrum of resistance mechanisms. In addition, the issue of false-positive results has to be seriously considered [
4]. Culture techniques are easier to perform, cheaper, and yield isolates for epidemiological follow-up.
With our classic manual workflow for screening samples (MRSA, MDRGN, and VRE), we had to read each plate twice and could only create final reports on day 1 if there was growth on MRSA or VRE plates. Negative reports could not be issued prior to day 2 (i.e., up to 48 hours after inoculation). This workflow was originally created to reliably fulfill the incubation times mentioned in the respective package inserts (18–48 hours) in a laboratory with working hours from 07:30 a.m. to 06:00 p.m. Using TLA, we wanted to establish a workflow with one single imaging time point within the first 24 hours of incubation that enabled final reports on day 1. This would substantially shorten the time-to-report and would halve the workload for laboratory technicians.
The incubation times needed for visible growth of bacteria were determined mainly based on rapid susceptibility testing studies. van den Bijllaardt et al. [
5] studied a total of 88 Enterobacteriaceae isolates and imaged agar diffusion plates every hour from one to 20 hours of incubation, concluding that all plates were readable after seven hours of incubation. Their findings support our results for MDRGN first growth and single colony appearance. Fröding et al. [
6] compared a six-hour reading time point of agar diffusion plates (
E. coli and
Klebsiella pneumoniae) with the standard 18-hour reading time point. They found that hetero-resistant isolates, in particular, were not correctly interpretable after six hours of incubation. Hombach et al. [
7] examined agar diffusion plates between six and 12 hours and compared results with the standardized reading after 18 hours, using
E. coli,
K. pneumoniae,
S. aureus, and
Staphylococcus epidermidis. They found that plates inoculated with
E. coli were reliably readable after six hours compared with plates inoculated with
S. aureus, which had to be read after 12 hours. Only one study, performed with streptococci, addressed the differentiation of
Streptococcus pneumoniae from other streptococci, using optochin susceptibility [
8]. That study demonstrated that an incubation time of 12 hours is best for reliable identification of pneumococci.
Because of the lack of growth kinetics on chromogenic media, we established our own kinetic readings in order to identify the optimal time points for imaging during patient sample processing. From sub-culturing procedures, we knew that sub-cultures of MRSA or MDRGN on blood agar plates showed first growth (depending on species) after approximately three to five hours of incubation. However, we did not know when first growth on selective chromogenic plates would become detectable. For the first 24–28 hours of incubation we chose a two-hour interval between individual images. This was short enough to gain an impression regarding growth and long enough to make clear decisions according to the following criteria for the three different time points: A) First growth: first growth was important to distinguish between negative reports (i.e., no growth) and cases in which follow-up work had to be done. If the chosen incubation times were too short, false negative reports would occur. B) Single colonies: single colonies were defined as the growth of colonies with which follow-up work could be directly performed, that is, without further sub-culturing. This time point is important for the laboratory routine because the right choice would reduce the need for sub-culturing, which would increase hands-on time and material costs. C) Typical morphology: typical morphology is an important time point because a direct conclusion can be drawn simply by looking at the morphology and color of the colonies on the plate.
The choice of species to evaluate the ESBL plate was based on screening procedure guidelines [
910].
P. aeruginosa and
A. baumannii complex are the only species of these genera that are relevant for screening in Germany. For the family Enterobacteriaceae,
E. coli and
K. pneumoniae are the most relevant species in terms of multi-resistance in Germany.
The earliest first growth was observed after four hours with gram-negative rods and high inoculum concentration (108/mL). The first MRSA were observed after six hours, and the first VRE appeared after eight hours. More importantly, single colonies appeared as early as 12, six, and 12 hours and as late as 18, 18, and 48 hours for MRSA, MDRGN, and VRE (VRESelect), respectively. Detection times on the other two VRE-plates used were not significantly shorter.
One main limitation of our study is that we did not evaluate earlier imaging time points (<20 hours) with patient samples. This was a deliberate decision as (1) an earlier imaging time point would not fall within the regular working hours of the laboratory, and (2) introducing an earlier imaging time point requiring a second read for each plate would contradict our goal of reducing workload.
Based on our results, we now use a 20-hour imaging time point for MRSA and MDRGN and an additional imaging time point at 36 hours only for VRE. In terms of workflow, this means that we read MRSA and MDRGN once after 20 hours of incubation. For VRE we have to wait for the 36-hour image to distinguish between growth and no growth. Therefore, we achieved our ultimate goal of reading plates once within the first 24 hours of incubation for two out of three multi-resistant bacteria and thus markedly reduced turn-around time from a median of 48 hours to a median of 24 hours. Additionally, we halved the reading workload for technicians. These findings would be enough evidence to implement shorter incubation times in all laboratories using the same agar plates as we did.
Go to :

Acknowledgment
We thank C. Becker and S. Faber for image reading and excellent technical assistance. We thank P. Steinke for selection of strains.
Go to :

Notes
Go to :

References
1. Rubinovitch B, Pittet D. Screening for methicillin-resistant
Staphylococcus aureus in the endemic hospital: what have we learned? J Hosp Infect. 2001; 47:9–18. PMID:
11161895.
2. Hagen RM, Seegmüller I, Navai J, Kappstein I, Lehn N, Miethke T. Development of a real-time PCR assay for rapid identification of methicillin-resistant
Staphylococcus aureus from clinical samples. Int J Med Microbiol. 2005; 295:77–86. PMID:
15969468.
3. Dalpke AH, Hofko M, Zimmermann S. Development of a real-time PCR protocol requiring minimal handling for detection of vancomycin-resistant enterococci with the fully automated BD Max system. J Clin Microbiol. 2016; 54:2321–2329. PMID:
27358466.
4. Blanc DS, Basset P, Nahimana-Tessemo I, Jaton K, Greub G, Zanetti G. High proportion of wrongly identified methicillin-resistant
Staphylococcus aureus carriers by use of a rapid commercial PCR assay due to presence of staphylococcal cassette chromosome element lacking the
mecA gene. J Clin Microbiol. 2011; 49:722–724. PMID:
21159945.
5. van den Bijllaardt W, Buiting AG, Mouton JW, Muller AE. Shortening the incubation time for antimicrobial susceptibility testing by disk diffusion for Enterobacteriaceae: how short can it be and are the results accurate? Int J Antimicrob Agents. 2017; 49:631–637. PMID:
28263895.
6. Fröding I, Vondracek M, Giske CG. Rapid EUCAST disc diffusion testing of MDR
Escherichia coli and
Klebsiella pneumoniae: inhibition zones for extended-spectrum cephalosporins can be reliably read after 6 h of incubation. J Antimicrob Chemother. 2017; 72:1094–1102. PMID:
27999046.
7. Hombach M, Jetter M, Blöchliger N, Kolesnik-Goldmann N, Böttger EC. Fully automated disc diffusion for rapid antibiotic susceptibility test results: a proof-of-principle study. J Antimicrob Chemother. 2017; 72:1659–1668. PMID:
28333189.
8. Burckhardt I, Panitz J, Burckhardt F, Zimmermann S. Identification of
Streptococcus pneumoniae: development of a standardized protocol for optochin susceptibility testing using total lab automation. Biomed Res Int. 2017; 2017:4174168. PMID:
28401155.
9. Hygiene measures for infection or colonization with multidrug-resistant gram-negative bacilli. Commission recommendation for hospital hygiene and infection prevention (KRINKO) at the Robert Koch Institute (RKI). Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2012; 55:1311–1354. PMID:
23011096.
10. Mattner F, Bange FC, Meyer E, Seifert H, Wichelhaus TA, Chaberny IF. Preventing the spread of multidrug-resistant gram-negative pathogens: recommendations of an expert panel of the German Society for Hygiene and Microbiology. Dtsch Arztebl Int. 2012; 109:39–45. PMID:
22334820.
Go to :










 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download