Abstract
Bronchiectasis is a chronic disease characterized by airway infection and inflammation, leading to permanent dilation of the bronchi. Evaluation of underlying etiology is important in managing young bronchiectasis patients with recurrent infections caused by unusual pathogens. The signal transducer and activator of transcription 1 (STAT1) protein plays a key role in STAT signaling and immune system regulation. Heterozygotes for gain-of-function (GOF) alleles of the STAT1 gene usually display autosomal dominant chronic mucocutaneous candidiasis (CMC) and a wide range of clinical features, such as bronchiectasis. Here, we report on a patient with CMC and bronchiectasis with various types of infections who carried a pathogenic variant of the STAT1 gene. The 24-year-old female presented with recurrent respiratory bacterial and nontuberculous mycobacterial infections complicated by severe bronchiectasis and CMC. Whole-exome sequencing revealed a c.800C>T (p.Ala267Val) heterozygous mutation in the STAT1 gene. Further analysis by Sanger sequencing of STAT1 from the patient and her parents revealed the patient had a de novo occurrence of the variant. This is the first report of a Korean patient with a GOF pathogenic variant in STAT1. Physicians should be aware of the existence of this variant as a genetic factor associated with CMC and bronchiectasis complicated by recurrent infection.
Bronchiectasis is a chronic respiratory disease characterized by airway infection and inflammation, leading to permanent dilation of the bronchi.123 Bronchiectasis may be a consequence of structural lung damage caused by prior respiratory infection or systemic disorders associated with genetic defects, such as cystic fibrosis. Thus, evaluating causative or correctable underlying factors is important in the management of patients with respiratory infections complicated by bronchiectasis.4
The signal transducer and activator of transcription 1 (STAT1) plays an important role in STAT signaling and immune system regulation.5 In heterozygotes for gain-of-function (GOF) alleles in the STAT1 gene, chronic mucocutaneous candidiasis (CMC) is common, whereas loss-of-function variants in STAT1 give rise to a broad range of diseases, including bacterial, viral, or nontuberculous mycobacterial (NTM) infection.678 In addition, recent studies have demonstrated an association between STAT1 GOF variants and a broader range of clinical phenotypes, including bronchiectasis and respiratory infections.910111213 Here, we report the first case of a young Korean female with a de novo STAT1 GOF pathogenic variant who presented with recurrent respiratory bacterial and NTM infection complicated by CMC and severe bronchiectasis.
This case report was approved by the Institutional Review Board of Samsung Medical Center (IRB No. 2018-07-140). Patient information was anonymized and de-identified, and informed consent was waived. Our patient experienced repeated episodes of pneumonia commencing in childhood, with bronchiectasis first documented at the age of 12 years. She has no history of tuberculosis. When the patient was 18 years old, she had pneumonia caused by Pseudomonas aeruginosa. At the age of 23 years, the patient started to suffer from NTM lung disease caused by macrolide-susceptible Mycobacterium massiliense; the patient responded poorly to antibiotic treatment, including clarithromycin, rifampin, ethambutol, and intravenous amikacin, for 10 months at another hospital and was transferred to our hospital.
The patients had oral thrush, as well as cutaneous candidiasis of nasal skin (Fig. 1A) and onychomycosis of a fingernail (Fig. 1B), with the diagnoses confirmed by the observation of fungal hyphae on periodic acid-Schiff staining on biopsy specimens. A chest computed tomography scan revealed bilateral severe bronchiectasis and bronchiolitis in both lungs (Fig. 1C). Sputum examinations revealed multiple positive cultures for P. aeruginosa and M. massiliense, and the M. massiliense isolate was still susceptible to macrolide. The patient was treated for CMC with itraconazole. Multidrug antibiotic therapy consisting of amikacin, azithromycin, clofazimine, and linezolid was administered for M. massiliense lung disease.
Further tests, including CFTR gene testing and quantitation of serum immunoglobulin (Ig) and alpha-1-antitrypsin, were performed to find the underlying etiology in this patient. However, we found no pathogenic variants of the CFTR gene, and the levels of serum IgG (1580 mg/dL), IgA (90 mg/dL), IgM (94 mg/dL), and alpha-1-antitrypsin (173.7 mg/dL) were normal. To rule out primary ciliary dyskinesia, electron microscopy of respiratory epithelial samples was performed, but showed normal cilia structures.
Due to uncertainty regarding the basic etiology, we performed whole-exome sequencing (WES), with the written informed consent of the patient, to identify genetic causes of the disease. Genomic DNA was extracted and captured with the Agilent SureSelect Human All Exon V5 Kit (Agilent Technologies, Santa Clara, CA, USA) and sequenced on the Illumina Next-Seq500 platform (Illumina Inc., San Diego, CA, USA). Raw sequence reads were processed and aligned to the hg19 human genome reference sequence. A mean coverage of 99.05× was achieved, and 95.4% of targeted bases were read >20 times by exome capture and sequencing. A total of 99028 variants were identified. After screening pneumonia-related or CMC-related genes, we identified a missense variant (NM_007315.3:c.800C>T, p.Ala267Val) in the coiled-coil domain of the STAT1 gene (Fig. 2A). This variant was not reported in the Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org) nor in the Korean Reference Genome database (KRGDB, http://152.99.75.168/KRGDB). According to the 2015 guidelines from the American College of Medical Genetics and Genomics and the Association of Molecular Pathology,14 this p.Ala267Val variant can be categorized as a “pathogenic” variant based on the following factors: PS1, same amino acid change as a previously established pathogenic variant, and PS3, well-established functional studies support its damaging effect on the gene product.6131415 Sanger sequencing confirmed the heterozygous STAT1 variant (NM_007315.3:c.800C>T, p.Ala267Val) in the patient, whereas her parents did not have the variant, indicating that the variant occurred de novo (Fig. 2B).
We present the first case of a Korean patient with a de novo STAT1 GOF variant. She presented with recurrent respiratory infection by unusual pathogens complicated by severe bronchiectasis and CMC. GOF variants of the STAT1 gene affect the coiled-coil domain of STAT1, increase STAT1 phosphorylation by impairing nuclear dephosphorylation, and eventually can cause diminished numbers of interleukin-17-producing T cells.7 The heterozygous c.800C>T (p.Ala267Val) variant in STAT1 identified in our patient was originally described as an autosomal dominant GOF pathogenic variant causing CMC. However, recent studies have demonstrated the potential association between these variants and more broad-ranging clinical phenotypes, including sinopulmonary disease and severe bronchiectasis.910111213 Although the underlying mechanisms remain elusive, it was suggested that degrees of impairment in B cells, especially class switched memory B cells, in patients with STAT1 GOF variants might contribute to the phenotypic variation in sinopulmonary manifestation.13 STAT1 GOF variants are also reported to be associated with various kinds of infectious disease, autoimmune disease, cerebral aneurysms, and cancer.91011121315161718 Regarding these topics, Toubiana, et al.9 recently reported detailed clinical spectrums in 274 patients from 40 countries with STAT11 GOF pathogenic variant: 98% of them had CMC, 37% had autoimmune manifestations, 21% had bronchiectasis, and 6% had mycobacterial infections, including tuberculosis and NTM infections.
In our case, diagnosis of underlying disease of bronchiectasis and recurrent infections led to the patient's and her parents' psychological stability and compliance to treatment, and it eventually suggested the possibility of application of targeted therapy, such as Janus kinase (JAK) family tyrosine kinase inhibitor, such as ruxolitinib, for disease control.19 Therefore, our case highlights the importance of a thorough evaluation of the underlying etiology in patients with bronchiectasis, particularly those with chronic recurrent infections by unusual pathogens.
In our case, we used WES to identify basic etiology and discovered the STAT1 GOF variant. The diagnosis of STAT1-related disorders based on clinical features can be hampered since patients with autosomal dominant STAT1 GOF variants often present broad clinical phenotypes. Therefore, patients with this rare genetic disease might undergo a futile and time-consuming process for a definitive diagnosis.13 WES can be a useful and cost-effective diagnostic tool, especially for patients with broad or unusual clinical presentation.
In summary, this report describes the first case of a Korean patient carrying a pathogenic variant of the STAT1 gene, which likely resulted in the patient's recurrent respiratory bacterial and NTM infection complicated by severe bronchiectasis and CMC. Our report highlights the need for physicians to be aware of GOF pathogenic variants in the STAT1 gene as a genetic factor associated with CMC and bronchiectasis complicated by infection by various pathogens.
Figures and Tables
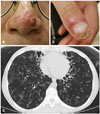 | Fig. 1Fungal infections and severe bronchiectasis in the 24-year-old female patient. (A) Cutaneous candidiasis of nasal skin. (B) Onychomycosis of a fingernail. (C) Bilateral bronchiectasis and bronchiolitis on chest computed tomography scan. |
 | Fig. 2Pathogenic STAT1 variant identified in the 24-year-old female patient. (A) Integrative Genomics Viewer snapshot of the STAT1 pathogenic variant (NM_007315.3:c.800C>T, p.Ala267Val) (arrow) identified by whole-exome sequencing. (B) Validation of the STAT1 variant by Sanger sequencing. Chromatograms show the heterozygous variant in the proband (patient), indicated by the arrow, and the normal sequence in the unaffected parents. |
ACKNOWLEDGEMENTS
This research was supported by a grant of the Korea Health technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI15C2778).
References
1. Polverino E, Goeminne PC, McDonnell MJ, Aliberti S, Marshall SE, Loebinger MR, et al. European Respiratory Society guidelines for the management of adult bronchiectasis. Eur Respir J. 2017; 50:1700629.

2. Chalmers JD, Aliberti S, Blasi F. Management of bronchiectasis in adults. Eur Respir J. 2015; 45:1446–1462.

3. Pasteur MC, Bilton D, Hill AT. British Thoracic Society Bronchiectasis non-CF Guideline Group. British Thoracic Society guideline for non-CF bronchiectasis. Thorax. 2010; 65:Suppl 1. i1–i58.

4. Araújo D, Shteinberg M, Aliberti S, Goeminne PC, Hill AT, Fardon T, et al. Standardised classification of the aetiology of bronchiectasis using an objective algorithm. Eur Respir J. 2017; 50:1701289.

5. Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, et al. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006; 2:e131.
6. van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011; 365:54–61.

7. Boisson-Dupuis S, Kong XF, Okada S, Cypowyj S, Puel A, Abel L, et al. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr Opin Immunol. 2012; 24:364–378.

8. Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, et al. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001; 293:300–303.

9. Toubiana J, Okada S, Hiller J, Oleastro M, Lagos Gomez M, Aldave Becerra JC, et al. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood. 2016; 127:3154–3164.

10. Depner M, Fuchs S, Raabe J, Frede N, Glocker C, Doffinger R, et al. The extended clinical phenotype of 26 patients with chronic mucocutaneous candidiasis due to gain-of-function mutations in STAT1. J Clin Immunol. 2016; 36:73–84.

11. Dotta L, Scomodon O, Padoan R, Timpano S, Plebani A, Soresina A, et al. Clinical and immunological data of nine patients with chronic mucocutaneous candidiasis disease. Data Brief. 2016; 7:311–315.

12. Dotta L, Scomodon O, Padoan R, Timpano S, Plebani A, Soresina A, et al. Clinical heterogeneity of dominant chronic mucocutaneous candidiasis disease: presenting as treatment-resistant candidiasis and chronic lung disease. Clin Immunol. 2016; 164:1–9.

13. Breuer O, Daum H, Cohen-Cymberknoh M, Unger S, Shoseyov D, Stepensky P, et al. Autosomal dominant gain of function STAT1 mutation and severe bronchiectasis. Respir Med. 2017; 126:39–45.

14. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17:405–424.

15. Zheng J, van de Veerdonk FL, Crossland KL, Smeekens SP, Chan CM, Al Shehri T, et al. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC). Eur J Immunol. 2015; 45:2834–2846.

16. Pedraza-Sánchez S, Lezana-Fernández JL, Gonzalez Y, Martínez-Robles L, Ventura-Ayala ML, Sadowinski-Pine S, et al. Disseminated tuberculosis and chronic mucocutaneous candidiasis in a patient with a gain-of-function mutation in signal transduction and activator of transcription 1. Front Immunol. 2017; 8:1651.

17. Koo S, Kejariwal D, Al-Shehri T, Dhar A, Lilic D. Oesophageal candidiasis and squamous cell cancer in patients with gain-of-function STAT1 gene mutation. United European Gastroenterol J. 2017; 5:625–631.

18. Sampaio EP, Hsu AP, Pechacek J, Bax HI, Dias DL, Paulson ML, et al. Signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations and disseminated coccidioidomycosis and histoplasmosis. J Allergy Clin Immunol. 2013; 131:1624–1634.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download