Abstract
Background
Acral melanomas are known to have a low frequency of BRAF mutation, in contrary to higher KIT mutation. Recently, VE1 immunostaining was reported to have a good correlation with BRAF mutation status.
Objective
We aimed to evaluate the clinicopathological features of BRAF-mutated acral melanomas and validate the correlation of the VE1 immunohistochemical stains in those cases.
Methods
The clinical features (age, sex, anatomical site), and histopathological characteristics of 41 patients with acral melanoma were evaluated. We performed a next-generation sequencing to detect BRAF mutation status. We also determined the correlation of VE1 immunohistochemical staining with BRAF mutation status.
Results
Among 19 acral melanomas with BRAF mutation, common histopathological subtype was acral lentiginous melanoma (8/19, 42%) and nodular melanoma (8/19, 42%) and superficial spreading melanoma (3/19, 16%) followed. VE1 immunostaining results were positive in all 15 cases with BRAF V600E mutation (sensitivity 100%), and negative in 4 cases of BRAF non-V600E mutation. However, VE1 immunostaining was negative in all 22 patients with BRAF wild-type.
Cutaneous melanoma is a fatal skin cancer, and its occurrence has increased dramatically in recent decades. Superficial spreading melanoma (SSM) and nodular melanoma (NM) are frequently observed in Caucasian populations, whereas acral lentiginous melanoma (ALM) is the most common type in Asian patients1. Recently, genetic mutations in genes such as BRAF, NRAS, KIT are frequently found in melanomas. Somatic oncogenic mutations of BRAF have emerged as both an effective biomarker and a therapeutic target, and two BRAF inhibitors, such as vemurafenib and dabrafenib, are Food and Drug Administration-approved for the treatment of unresectable or metastatic melanoma23456. Among them, a single amino acid substitution from valine to glutamic acid at codon 600 occurs in 90% of mutated cases. Therefore, the detection of BRAF mutation in melanoma is crucial for patient treatment. SSM tends to have a high frequency of BRAF mutations at about 75%, whereas acral melanomas on the palms, soles, and nails are known to have a low frequency of BRAF mutations7. Most of studies from Caucasian population did not include acral melanomas, and there have been only a few studies on BRAF mutations and acral melanomas in Asian patients7891011. Recently, a monoclonal antibody against mutant BRAF V600E protein (VE1) has been developed1213141516. Several studies have indicated that immunohistochemical (IHC) staining with VE1 can be used to accurately detect the BRAF V600E mutation in brain metastases, papillary thyroid carcinoma, malignant melanoma, lung adenocarcinoma, and serous ovarian tumor17. Furthermore, many studies have validated the sensitivity and specificity of IHC stains in melanoma tissues1213141516. This study aimed to evaluate the clinicopathological features of BRAF mutations in acral melanomas in Korean patients and to assess the reliability of VE1 IHC staining compared with genetic analysis for detection of BRAF mutation.
A total of 41 patients with acral melanoma were enrolled from the Department of Dermatology at Chonnam National University Hospital in Gwangju and Chonnam National University Hwasun Hospital in Hwasun, South Korea from January 2004 to June 2017. The study protocol was approved by the institutional review board of Chonnam National University Hwasun Hospital (IRB no. CNUHH-2016-008). We received the patient's consent form about publishing all photographic materials and the study was conducted according to the Declaration of Helsinki Principles. We retrospectively reviewed the clinical characteristics such as age, sex, location, and histopathological features.
Primary cutaneous melanoma tissue samples were obtained via surgical or punch biopsies. BRAF mutation testing was performed on sections from archival formalin-fixed paraffin-embedded (FFPE) tissue blocks of acral melanomas. Specimens were sectioned with 10 µm thickness from FFPE blocks and put on the slides. Tumor presence was verified by hematoxylin and eosin (H&E) staining. Areas containing viable tumors were marked on the H&E slides. In comparison with non-tumor tissue components, dissected areas contained a minimum of 50% tumor nuclei. Paraffin was removed by xylene treatment, and DNA was purified using the QIAamp DNA FFPE Tissue KIT (QIAGEN GmbH, Hilden, Germany)18.
A targeted panel was used to capture the target region covering the BRAF gene, which included all the coding exons of the gene for the detection of single nucleotide variants (SNVs), and insertions/deletions (INDELs). Genomic DNA (200~500 ng) was prepared to construct libraries with the SureSelect targeted panel according to manufacturer's protocol. Briefly, the qualified genomic DNA sample was randomly fragmented by Covaris followed by adapter ligation, purification, hybridization, and polymerase chain reaction. Captured libraries were subjected to Agilent 2100 Bioanalyzer to estimate the quality and were loaded onto the Illumina HiSeq 2500 (TheragenEtex Bio Institute, Suwon, Korea) according to the manufacturer's recommendations. Raw image files were processed by HCS1.4.8 for base-calling with default parameters, and the sequences of each individual were generated as 101 bp paired-end reads.
At the NGS data pre-processing step, sequence reads were aligned to the human genome (hg19) using BWA-MEM19. For analysis-ready BAM file generation, the overall pre-processing procedure, including removal of duplication, local realignment, and recalibration, was performed using GATK Best Practices of the Broad Institute.20
At the variant discovery step, SNV and INDEL were analyzed using three open source callers (UnifiedGenotyper21, Freq22, SNVer23, and Samsung SDS's in-house callers). SNVs and INDELs were filtered using germline mutations and false positive filters. SNVs with a variant allele frequency of ≥5% and INDELs of ≥10% were selected as the final result.
We reviewed all histopathological slides of the 41 patients and compared their features according to BRAF mutation status. We determined the histopathological subtypes of acral melanomas (SSM, ALM, NM, or not characterized).
IHC staining for 41 acral melanoma specimens was performed using the mouse anti-human BRAF monoclonal antibody (Spring Bioscience, Pleasanton, CA, USA). IHC analysis was performed on 4-µm paraffin-embedded sections following standard procedures. Antigen retrieval was performed with Tris/EDTA buffer solution (pH 9) (Dako, Carpinteria, CA, USA), and primary antibody incubation was performed for 1 hour and 30 minutes at room temperature using the BRAF V600E mutation-specific antibody VE1 (Spring Biosciences, Pleasanton, CA, USA) diluted at 1:100. IHC stains were visualized using a polymer-based system (EnVision; Dako) with 3-amino-9-ethylcarbazole (AEC) as the chromogen. Positive controls (ameloblastoma with known BRAF V600E mutation) and negative controls (inflammatory fibrous hyperplasia and omission of the primary antibody) were included in each reaction. One dermatologist and one pathologist reviewed the IHC stained slides. All observers were unaware of the molecular results. Immunoreactions were analyzed based on the degree of cytoplasmic immunostaining (0~3+). Negative cytoplasmic staining was scored as 0 (negative), whereas positive cytoplasmic staining was scored as 1+ (mild intensity), 2+ (moderate intensity), or 3+ (strong intensity).
Of 19 acral melanoma patients with BRAF mutation, there were 6 male patients and 13 female patients, with a mean age of 58±17 years (Table 1). In terms of melanoma location on the acral sites, there were 3 acral melanomas on the thumbnails, one on the finger, one on the great toenail, one on the toe, and 13 on the sole (Fig. 1A, C). The most common histopathological subtype was ALM (8/19, 42%) and NM (8/19, 42%) and SSM (3/19, 16%) followed (Fig. 1B, D).
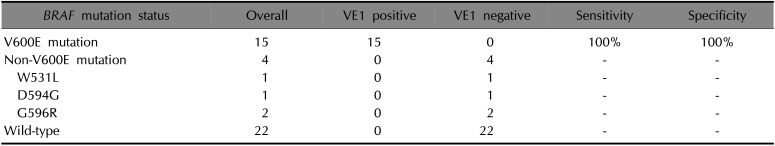
According to NGS, 19 cases were identified as having BRAF mutation, consisting of BRAF V600E (n=15), W531L (n=1), D594G (n=1), and G596R (n=2) (Table 1). A total 19 BRAF-mutated acral melanomas were analyzed with VE1 immunostaining. The results revealed a positive reaction in 15 acral melanoma cases with BRAF V600E mutation (Fig. 2), whereas 4 cases with BRAF non-V600E mutation were negative. Among BRAF-mutated melanomas, VE1 staining demonstrated a higher correlation with the V600E mutation than with non-V600E mutations. All 22 patients with BRAF wild type showed negative to VE1 immunostaining (Table 2). Therefore, VE1 stain showed 100% sensitivity and 100% specificity to BRAF V600E mutation.
In this study, we detected BRAF mutation in primary acral melanomas by both molecular study and easily-done immunohistochemical study. The rapid development of tools for genetic studies, such as NGS and genome-wide association study, has made it possible to accurately detect genetic mutations with high sensitivity from a relatively small amount of DNA, compared with Sanger sequencing and reverse transcription polymerase chain reaction3. BRAF mutations in acral melanomas in Asian patients may be underestimated due to difference of diagnostic methods. We performed NGS in this study and detected a higher frequency of BRAF mutations in acral melanomas. Most of BRAF mutation studies were performed in Caucasian populations in which the histopathological subtype of SSM is the most common. On the other hand, ALM is known to be the most common subtype in acral melanomas in which the BRAF mutation is relatively rare. In this study, we found that ALM and NM were more common than SSM in BRAF-mutated acral melanomas.
As BRAF inhibition is the reference treatment of BRAF V600E mutant metastatic melanomas, screening for BRAF mutations is necessary for an effective treatment with targeted therapies23456. Even though BRAF V600E mutation is the major mutation site, BRAF non-V600E mutation, such as V600K, and different codon sites of BRAF, were also found24. Recently, in Asian populations, BRAF non-V600E mutations also reported, and patients with BRAF D594/G596 mutation showed significantly better prognosis than patients with BRAF V600E and wild-type25. In our study, we also found that BRAF non-V600E, such as D594G, G596R, and W531L, however, there were no BRAF V600K, V600R, and V500Q mutations.
In this study, we demonstrated a high sensitivity of 79% and a specificity of 100% for the BRAF mutation with VE1 immunostaining, which had a good correlation with BRAF V600E mutation status with 100% sensitivity and specificity. In addition, all 22 patients with BRAF wild-type revealed negative results to VE1 immunostain. There were no false-positive or false-negative cases.
To validate IHC screening, the staining intensity must be strong enough to be distinguished from an artifact or melanin pigmentation. In our case, the detection of red intensity was more noticeable. As the differentiation between a tumor cell and a melanophage was difficult, 3,3′-diaminobenzidine-detected immunostained tumor cells could not be identified and AEC-detected immunostained tumor cells solved the ambiguity of stained images. We acknowledge that there are a number of limitations in our study. Firstly, our study was a single-center study with a limited number of patients. Another limitation was that this study was unclear on how to manage samples with unclear staining. In the study by Boursault et al.26, 3 cases remained unclear because wild-type primary melanoma showed a faint brown staining. Rapisuwon et al.27 also reported cases of weak (1+) VE1-stained BRAF mutant specimens but determined these as positive IHC stains. On the contrary, a few reports regarded a weak staining as negative2829. These differences point out the need for definitive technical interpretive criteria in mutation-specific IHC staining. In our study, among 15 BRAF V600E mutation, VE1 immunostain with 1+ reaction was observed in 3 patients, even though 12 patients showed moderate to strong intensity. Therefore, it would be needed to improve immunohistochemical technique and interpretation to decrease weak positive cases. Although both molecular and IHC analysis may have to be taken into conclusion when determining BRAF status, the main advantage of IHC testing is the accessibility of the test that can be accomplished in the same time as histopathological examination and used for patients with insufficient melanoma tissue for genetic analysis.
In conclusion, we detected BRAF mutations in acral melanomas. NGS is an improved and promising new technology for detecting genetic mutations. However, VE1 immunostaining had a good correlation with BRAF V600E mutation status in acral melanoma. These results suggest the strong analytical potential of the relationship between BRAF V600E mutations and VE1 immunohistochemical staining in acral melanomas, which would be a more powerful approach for managing individual patients and guiding research and treatment.
ACKNOWLEDGMENT
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1D1A3B03930554).
References
2. National Cancer Institute. FDA approval for vemurafenib [Internet]. Rockville, MD: National Cancer Institute [cited 2011 Sep 9];Available from: http://cancer.gov/about-cancer/treatment/drugs/fda-vemurafenib.
3. Wilson MA, Nathanson KL. Molecular testing in melanoma. Cancer J. 2012; 18:117–123. PMID: 22453011.

4. National Cancer Institute. FDA approval for Dabrafenib [Internet]. Rockville, MD: National Cancer Institute;cited 2013 Jun 21. Available from: https://www.cancer.gov/aboutcancer/treatment/drugs/dabrafenib.
5. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002; 417:949–954. PMID: 12068308.

6. Rubinstein JC, Sznol M, Pavlick AC, Arian S, Cheng E, Bacchiocchi A, et al. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J Transl Med. 2010; 8:67. PMID: 20630094.

7. Edlundh-Rose E, Egyházi S, Omholt K, Månsson-Brahme E, Platz A, Hansson J, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006; 16:471–478. PMID: 17119447.

8. Saldanha G, Potter L, Daforno P, Pringle JH. Cutaneous melanoma subtypes show different BRAF and NRAS mutation frequencies. Clin Cancer Res. 2006; 12:4499–4505. PMID: 16899595.

9. Lang J, MacKie RM. Prevalence of exon 15 BRAF mutations in primary melanoma of the superficial spreading, nodular, acral, and lentigo maligna subtypes. J Invest Dermatol. 2005; 125:575–579. PMID: 16117801.

10. Jin SA, Chun SM, Choi YD, Kweon SS, Jung ST, Shim HJ, et al. BRAF mutations and KIT aberrations and their clinicopathological correlation in 202 Korean melanomas. J Invest Dermatol. 2013; 133:579–582. PMID: 23014346.

11. Hong JW, Lee S, Kim DC, Kim KH, Song KH. Prognostic and clinicopathologic associations of BRAF mutation in primary acral lentiginous melanoma in Korean patients: a preliminary study. Ann Dermatol. 2014; 26:195–202. PMID: 24882974.
12. Pearlstein MV, Zedek DC, Ollila DW, Treece A, Gulley ML, Groben PA, et al. Validation of the VE1 immunostain for the BRAF V600E mutation in melanoma. J Cutan Pathol. 2014; 41:724–732. PMID: 24917033.
13. Marin C, Beauchet A, Capper D, Zimmermann U, Julié C, Ilie M, et al. Detection of BRAF p.V600E mutations in melanoma by immunohistochemistry has a good interobserver reproducibility. Arch Pathol Lab Med. 2014; 138:71–75. PMID: 23651150.
14. Capper D, Preusser M, Habel A, Sahm F, Ackermann U, Schindler G, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011; 122:11–19. PMID: 21638088.

15. Long GV, Wilmott JS, Capper D, Preusser M, Zhang YE, Thompson JF, et al. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am J Surg Pathol. 2013; 37:61–65. PMID: 23026937.

16. Manfredi L, Meyer N, Tournier E, Grand D, Uro-Coste E, Rochaix P, et al. Highly concordant results between immunohistochemistry and molecular testing of mutated V600E BRAF in primary and metastatic melanoma. Acta Derm Venereol. 2016; 96:630–634. PMID: 26695089.

17. Capper D, Berghoff AS, Magerle M, Ilhan A, Wöhrer A, Hackl M, et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012; 123:223–233. PMID: 22012135.

18. Yaman B, Kandiloğlu G, Akalin T. BRAF-V600 Mutation heterogeneity in primary and metastatic melanoma: a study with pyrosequencing and immunohistochemistry. Am J Dermatopathol. 2016; 38:113–120. PMID: 26630683.
19. Li H, Drubin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. PMID: 19451168.

20. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2010; 43:491–498.

21. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 43:11.10.1–11.10.33. PMID: 25431634.
22. Wilm A, Aw PP, Bertrand D, Yeo GH, Ong SH, Wong CH, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012; 40:11189–11201. PMID: 23066108.

23. Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011; 39:e132. PMID: 21813454.

24. Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017; 545:175–180. PMID: 28467829.

25. Wu X, Yan J, Dai J, Ma M, Tang H, Yu J, et al. Mutations in BRAF codons 594 and 596 predict good prognosis in melanoma. Oncol Lett. 2017; 14:3601–3605. PMID: 28927118.

26. Boursault L, Haddad V, Vergier B, Cappellen D, Verdon S, Bellocq JP, et al. Tumor homogeneity between primary and metastatic sites for BRAF status in metastatic melanoma determined by immunohistochemical and molecular testing. PLoS One. 2013; 8:e70826. PMID: 23976959.

27. Rapisuwon S, Busam KJ, Parks K, Chapman PB, Lee E, Atkins MB. Discordance between cobas BRAF V600 testing and VE1 immunohistochemistry in a melanoma patient with bone marrow metastases. Am J Dermatopathol. 2016; 38:687–689. PMID: 27541170.

28. Heinzerling L, Kühnapfel S, Meckbach D, Baiter M, Kaempgen E, Keikavoussi P, et al. Rare BRAF mutations in melanoma patients: implications for molecular testing in clinical practice. Br J Cancer. 2013; 108:2164–2171. PMID: 23579220.

29. Ihle MA, Fassunke J, König K, Grünewald I, Schlaak M, Kreuzberg N, et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014; 14:13. PMID: 24410877.

Fig. 1
Clinicopathological features of acral melanomas with BRAF V600E mutation. Acral melanomas (A) on the toe, (B) composed of pagetoid scatter and nests of epithelioid cells (H&E, ×100), and (C) on the finger, (D) showing heavily pigmented epithelioid cells (H&E, ×100).
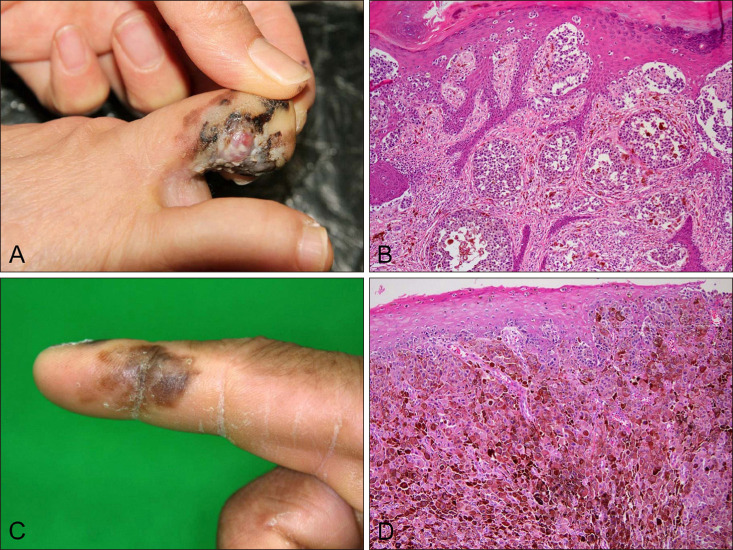
Fig. 2
VE1 immunohistochemical staining with acral melanomas with BRAF mutation. (A) Negative for VE1 immunostain in a case with BRAF wild-type (Grade 0, ×100). (B) Weak positive staining case with BRAF V600E mutation (Grade 1+, ×100). (C) Moderate positive staining case with BRAF V600E mutation (Grade 2+, ×100). (D) Strong positive staining case with BRAF V600E mutation (Grade 3+, ×100).
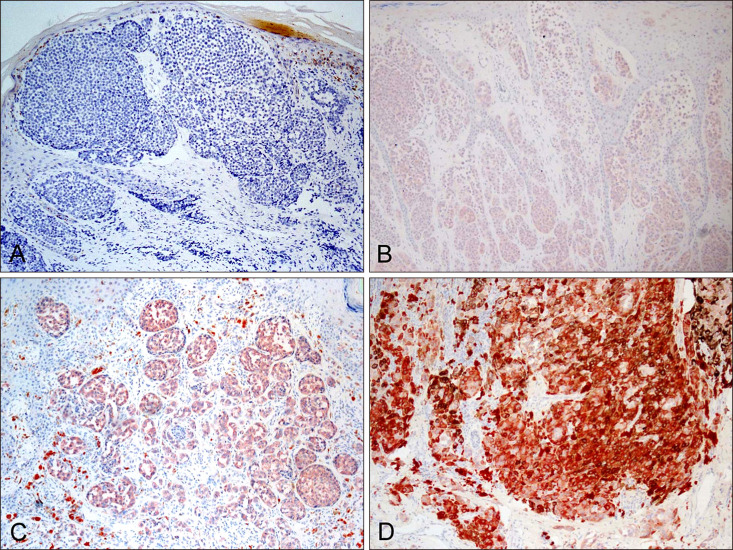
Table 1
Nineteen cases with VE1 immunostain grades according to BRAF mutation
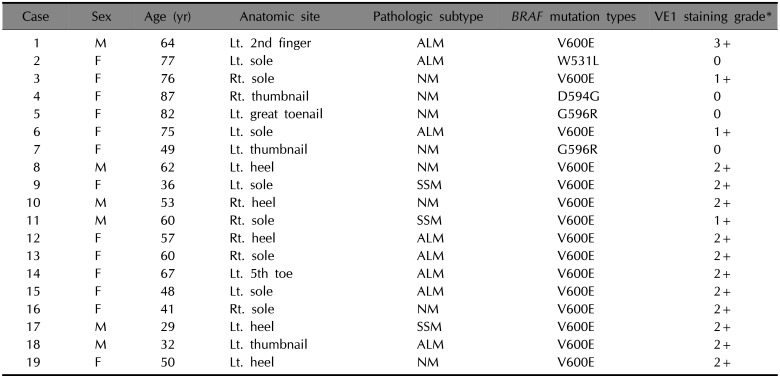
M: male, F: female, Lt.: left, Rt.: right, ALM: acral lentiginous melanoma, NM: nodular melanoma, SSM: superficial spreading melanoma, V: Valine, E: Glutamic acid, W: Tryptophan, L: Leucine, D: Aspartic acid, G: Glycine, R: Arginine. *0: negative, 1+: mild intensity, 2+: moderate intensity, 3+: strong intensity.
Table 2
Correlation of 41 acral melanomas with VE1 immunohistochemical stains with BRAF mutation status




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download