1. Xu D, Yuan R, Gu H, Liu T, Tu Y, Yang Z, et al. The effect of ultraviolet radiation on the transforming growth factor beta 1/Smads pathway and p53 in actinic keratosis and normal skin. Arch Dermatol Res. 2013; 305:777–786. PMID:
23632819.

2. Renzi C, Mastroeni S, Mannooranparampil TJ, Passarelli F, Caggiati A, Pasquini P. Skin cancer knowledge and preventive behaviors among patients with a recent history of cutaneous squamous cell carcinoma. Dermatology. 2008; 217:74–80. PMID:
18424897.

3. Ra SH, Li X, Binder S. Molecular discrimination of cutaneous squamous cell carcinoma from actinic keratosis and normal skin. Mod Pathol. 2011; 24:963–973. PMID:
21743436.

4. Anwar J, Wrone DA, Kimyai-Asadi A, Alam M. The development of actinic keratosis into invasive squamous cell carcinoma: evidence and evolving classification schemes. Clin Dermatol. 2004; 22:189–196. PMID:
15262304.

5. Berner A. [Actinic keratosis and development of cutaneous squamous cell carcinoma]. Tidsskr Nor Laegeforen. 2005; 125:1653–1654. Norwegian. PMID:
15976832.
6. Mittelbronn MA, Mullins DL, Ramos-Caro FA, Flowers FP. Frequency of pre-existing actinic keratosis in cutaneous squamous cell carcinoma. Int J Dermatol. 1998; 37:677–681. PMID:
9762818.

7. Kitagishi Y, Matsuda S, Minami A, Ono Y, Nakanishi A, Ogura Y. Regulation in cell cycle via p53 and PTEN tumor suppressors. Cancer Stud Mol Med Open J. 2014; 1:1–7.
8. Neto PD, Alchorne M, Michalany NS, Abreu M, Borra R. Reduced P53 staining in actinic keratosis is associated with squamous cell carcinoma: a preliminary study. Indian J Dermatol. 2013; 58:325.

9. Kanellou P, Zaravinos A, Zioga M, Stratigos A, Baritaki S, Soufla G, et al. Genomic instability, mutations and expression analysis of the tumour suppressor genes p14(ARF), p15(INK4b), p16(INK4a) and p53 in actinic keratosis. Cancer Lett. 2008; 264:145–161. PMID:
18331779.

10. Li-Weber M, Treiber MK, Giaisi M, Palfi K, Stephan N, Parg S, et al. Ultraviolet irradiation suppresses T cell activation via blocking TCR-mediated ERK and NF-kappa B signaling pathways. J Immunol. 2005; 175:2132–2143. PMID:
16081779.
11. Lee YB, Kim EK, Park HJ, Cho BK, Park YM, Kim JW, et al. Expression of Fas and Fas ligand in primary cutaneous squamous cell carcinoma in association with grade of tumor differentiation. Int J Dermatol. 2013; 52:1092–1097. PMID:
23045958.

12. Hameetman L, Commandeur S, Bavinck JN, Wisgerhof HC, de Gruijl FR, Willemze R, et al. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer. 2013; 13:58. PMID:
23379751.

13. Padilla RS, Sebastian S, Jiang Z, Nindl I, Larson R. Gene expression patterns of normal human skin, actinic keratosis, and squamous cell carcinoma: a spectrum of disease progression. Arch Dermatol. 2010; 146:288–293. PMID:
20231500.

14. Nindl I, Dang C, Forschner T, Kuban RJ, Meyer T, Sterry W, et al. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol Cancer. 2006; 5:30. PMID:
16893473.
15. Lambert SR, Mladkova N, Gulati A, Hamoudi R, Purdie K, Cerio R, et al. Key differences identified between actinic keratosis and cutaneous squamous cell carcinoma by transcriptome profiling. Br J Cancer. 2014; 110:520–529. PMID:
24335922.

16. S Al-MurraniX GaoJ LeeS MalladiNZ Frantz. Hill's Pet Nutrition, Inc.Methods for the diagnosis, control and prophylaxis of inflammation and mitigation of inflammatory conditions in felines. United States patent. EP 25,688,253. 2016. 4. 27.
17. Tusher VG, Tibshirani R, Chu G. SAM (significance analysis of microarrays): permits statistical distinction between microarray signals. In : Rédei GP, editor. Encyclopedia of genetics, genomics, proteomics, and informatics. 3rd ed. Dordrecht: Springer;2008. p. 1755.
18. Nicol NH. Actinic keratosis: preventable and treatable like other precancerous and cancerous skin lesions. Plast Surg Nurs. 1989; 9:49–55. PMID:
2675146.
19. Cheng SY, Luk NM, Chong LY. Special features of non-melanoma skin cancer in Hong Kong Chinese patients: 10-year retrospective study. Hong Kong Med J. 2001; 7:22–28. PMID:
11406672.
20. Monroig O, Rotllant J, Cerdá-Reverter JM, Dick JR, Figueras A, Tocher DR. Expression and role of Elovl4 elongases in biosynthesis of very long-chain fatty acids during zebrafish Danio rerio early embryonic development. Biochim Biophys Acta. 2010; 1801:1145–1154. PMID:
20601113.

21. Harris RB, Foote JA, Hakim IA, Bronson DL, Alberts DS. Fatty acid composition of red blood cell membranes and risk of squamous cell carcinoma of the skin. Cancer Epidemiol Biomarkers Prev. 2005; 14:906–912. PMID:
15824162.

22. Watanabe R, Fujii H, Yamamoto A, Hashimoto T, Kameda K, Ito M, et al. Immunohistochemical distribution of cutaneous fatty acid-binding protein in human skin. J Dermatol Sci. 1997; 16:17–22. PMID:
9438903.

23. Jones SJ, Dicker AJ, Dahler AL, Saunders NA. E2F as a regulator of keratinocyte proliferation: implications for skin tumor development. J Invest Dermatol. 1997; 109:187–193. PMID:
9242506.

25. Seo H, Choi Y, Shim J, Yoo I, Ka H. Comprehensive analysis of prostaglandin metabolic enzyme expression during pregnancy and the characterization of AKR1B1 as a prostaglandin F synthase at the maternal-conceptus interface in pigs. Biol Reprod. 2001; 90:99.

26. Okano M, Yamamoto H, Ohkuma H, Kano Y, Kim H, Nishikawa S, et al. Significance of INHBA expression in human colorectal cancer. Oncol Rep. 2013; 30:2903–2908. PMID:
24085226.

27. XU D, Yuan RH, Chu Y, Tang Y, Gu H, Zhang L, et al. Regulation of p53 expression though transforming growth factor β1 (TGFβ1)/Smads pathway in actinic keratosis. J Dermatol. 2012; 45:638–640.
28. Li J, Zhang Y, Romagosa R, Saghari S, Elgart G, Miner JH, et al. Laminin 10 in the angiogenesis and invasion of squamous cell carcinoma. J Invest Dermatol. 2006; 124(Suppl):A21.
29. Mostafa WZ, WZ , Mahfouz SM, Bosseila M, Sobhi RM, Zaki NS. An Immunohistochemical study of laminin in cutaneous and mucosal squamous cell carcinomas. J Egypt Wom Dermatol Soc. 2007; 4:24–33.
30. Sun Y, Tu Y, He LI, Ji C, Cheng BO. High mobility group box 1 regulates tumor metastasis in cutaneous squamous cell carcinoma via the PI3K/AKT and MAPK signaling pathways. Oncol Lett. 2016; 11:59–62. PMID:
26870167.

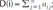
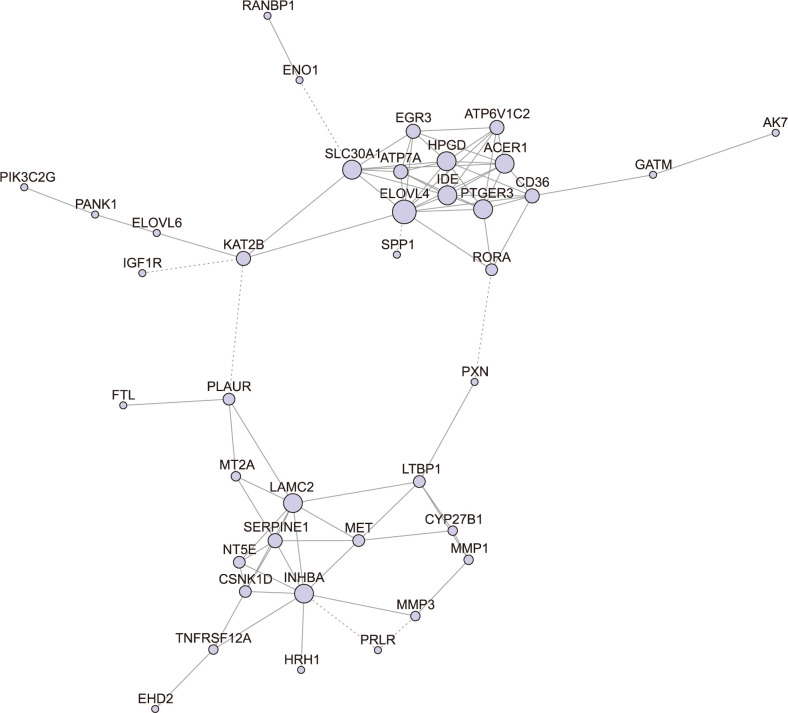
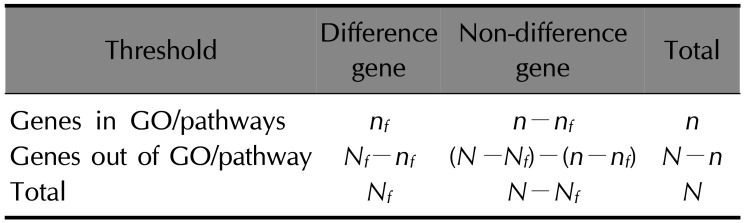
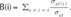 . Thereinto, σst represented total number of shortest path between s and t, while σst(i) represented the total number of the shortest path through i.
. Thereinto, σst represented total number of shortest path between s and t, while σst(i) represented the total number of the shortest path through i.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



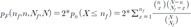
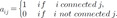
 XML Download
XML Download