1. Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann Nutr Metab. 2015; 66(Suppl 1):8–16. PMID:
25925336.

2. Bjerg A, Hedman L, Perzanowski MS, Platts-Mills T, Lundbäck B, Rönmark E. Family history of asthma and atopy: in-depth analyses of the impact on asthma and wheeze in 7- to 8-year-old children. Pediatrics. 2007; 120:741–748. PMID:
17908760.

3. Bantz SK, Zhu Z, Zheng T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. 2014; 5:pii: 202.

4. Li MX, Kwan JS, Bao SY, Yang W, Ho SL, Song YQ, et al. Predicting mendelian disease-causing non-synonymous single nucleotide variants in exome sequencing studies. PLoS Genet. 2013; 9:e1003143. PMID:
23341771.

5. Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007; 80:727–739. PMID:
17357078.

6. Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 2014; 15:256–278. PMID:
23341494.

7. Yoo J, Omori M, Gyarmati D, Zhou B, Aye T, Brewer A, et al. Spontaneous atopic dermatitis in mice expressing an inducible thymic stromal lymphopoietin transgene specifically in the skin. J Exp Med. 2005; 202:541–549. PMID:
16103410.

8. Zhu Z, Oh MH, Yu J, Liu YJ, Zheng T. The role of TSLP in IL-13-induced atopic march. Sci Rep. 2011; 1:23. PMID:
22355542.

9. Han H, Roan F, Ziegler SF. The atopic march: current insights into skin barrier dysfunction and epithelial cell-derived cytokines. Immunol Rev. 2017; 278:116–130. PMID:
28658558.

10. Ziegler SF. The role of thymic stromal lymphopoietin (TSLP) in allergic disorders. Curr Opin Immunol. 2010; 22:795–799. PMID:
21109412.

11. Leyva-Castillo JM, Hener P, Jiang H, Li M. TSLP produced by keratinocytes promotes allergen sensitization through skin and thereby triggers atopic march in mice. J Invest Dermatol. 2013; 133:154–163. PMID:
22832486.

12. Harada M, Hirota T, Jodo AI, Doi S, Kameda M, Fujita K, et al. Functional analysis of the thymic stromal lymphopoietin variants in human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2009; 40:368–374. PMID:
18787178.

13. Garmhausen D, Hagemann T, Bieber T, Dimitriou I, Fimmers R, Diepgen T, et al. Characterization of different courses of atopic dermatitis in adolescent and adult patients. Allergy. 2013; 68:498–506. PMID:
23452057.

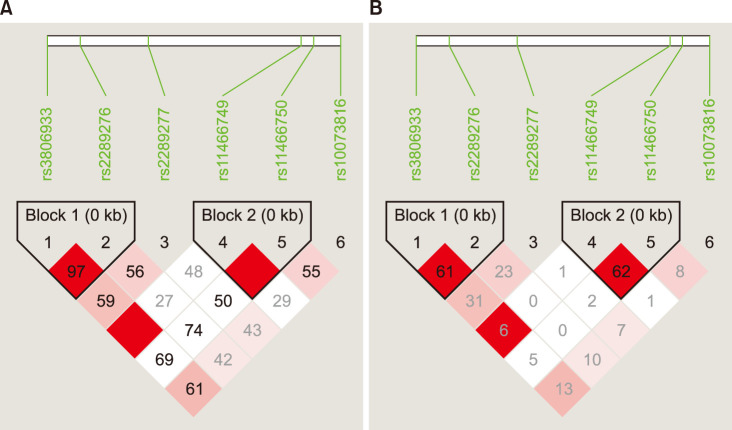
14. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21:263–265. PMID:
15297300.

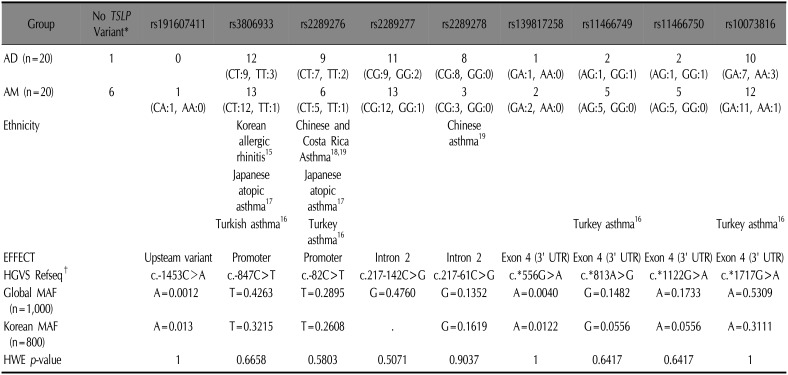
15. Zhang MH, Kim YS, Jin EH, Kim KM, Lee JH, Li CS, et al. Identification of polymorphisms in thymic stromal lymphopoietin (TSLP) gene and their association with allergic rhinitis. Genes Genomics. 2008; 30:461–468.
16. Birben E, Sahiner UM, Karaaslan C, Yavuz TS, Cosgun E, Kalayci O, et al. The genetic variants of thymic stromal lymphopoietin protein in children with asthma and allergic rhinitis. Int Arch Allergy Immunol. 2014; 163:185–192. PMID:
24525665.

17. Harada M, Hirota T, Jodo AI, Hitomi Y, Sakashita M, Tsunoda T, et al. Thymic stromal lymphopoietin gene promoter polymorphisms are associated with susceptibility to bronchial asthma. Am J Respir Cell Mol Biol. 2011; 44:787–793. PMID:
20656951.

18. Hunninghake GM, Soto-Quirós ME, Avila L, Kim HP, Lasky-Su J, Rafaels N, et al. TSLP polymorphisms are associated with asthma in a sex-specific fashion. Allergy. 2010; 65:1566–1575. PMID:
20560908.

19. Liu L, Xu LS, Liu QJ, Dong FZ, Qiu RF, Wen MC, et al. Two single nucleotide polymorphisms in TSLP gene are associated with asthma susceptibility in Chinese Han population. Exp Lung Res. 2012; 38:375–382. PMID:
22913730.

20. Irvine AD, McLean WH, Leung DY. Filaggrin mutations associated with skin and allergic diseases. N Engl J Med. 2011; 365:1315–1327. PMID:
21991953.

21. Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 2012; 69:3613–3634. PMID:
22538991.





 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download