Abstract
The tumor microenvironment greatly influences cancer cell characteristics, and acidic extracellular pH has been implicated as an essential factor in tumor malignancy and the induction of drug resistance. Here, we examined the characteristics of gastric carcinoma (GC) cells under conditions of extracellular acidity and attempted to identify a means of enhancing treatment efficacy. Acidic conditions caused several changes in GC cells adversely affecting chemotherapeutic treatment. Extracellular acidity did inhibit GC cell growth by inducing cell cycle arrest, but did not induce cell death at pH values down to 6.2, which was consistent with down-regulated cyclin D1 and up-regulated p21 mRNA expression. Additionally, an acidic environment altered the expression of atg5, HSPA1B, collagen XIII, collagen XXAI, slug, snail, and zeb1 genes which are related to regulation of cell resistance to cytotoxicity and malignancy, and as expected, resulted in increased resistance of cells to multiple chemotherapeutic drugs including etoposide, doxorubicin, daunorubicin, cisplatin, oxaliplatin and 5-FU. Interestingly, however, acidic environment dramatically sensitized GC cells to apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). Consistently, the acidity at pH 6.5 increased mRNA levels of DR4 and DR5 genes, and also elevated protein expression of both death receptors as detected by immunoblotting. Gene silencing analysis showed that of these two receptors, the major role in this effect was played by DR5. Therefore, these results suggest that extracellular acidity can sensitize TRAIL-mediated apoptosis at least partially via DR5 in GCs while it confers resistance to various type of chemotherapeutic drugs.
Tumor tissues are often exposed to acidic extracellular pH since tumors produce high levels of lactic acid and H+ by using the anaerobic glycolytic pathway for energy production [1]. Such extracellular acidification constitutes a stress to cells, and certain mechanisms by which tumor cells adapt to this stressful environment may enhance their malignant characteristics. Accumulating evidence has revealed that extracellular acidity increases the invasive behavior of cancer cells, including human malignant glioma and human melanoma cells, and promotes their survival by enabling evasion of apoptosis, consequently hindering tumor treatment [23].
Despite the development of multiple drug combination protocols and new treatment strategies, chemoresistance remains a major obstacle in cancer treatment [4]. Since the discovery that environmental acidity is a critical contributor to elevated drug resistance, various efforts have been and are currently being made to overcome acidity-induced chemoresistance such as the use of proton-pump inhibitors that block cellular proton release into the extracellular milieu [567].
Gastric cancer remains one of the leading causes of cancer-related death in several countries in Asia, Latin America, and Central and Eastern Europe [89]. Although surgical resection is effective in the early stage of gastric carcinoma (GC), additional treatment is necessary for advanced cases. However, despite considerable improvements in therapeutic regimens, the effectiveness of treatment and its benefit to survival have not significantly increased.
Considering that the stomach is frequently subjected to low pH due to the release of gastric acids, the diminished effectiveness of chemotherapy for gastric cancer may be related to the physiologically acidic environment found in this organ. It has recently been noted that an extremely acidic environment due to excessive secretion of gastric acids is often associated with the development of gastric cancer. Furthermore, gastric hyperacidity has been shown to promote GC cell invasion and induce chemoresistance [71011]. These results suggest that the characteristics of GC could be greatly influenced by acidic environments. However, relatively little study has been conducted concerning a means to overcome the deleterious acidic effects. As a part of study to improve the benefits of GC treatment under acidic condition, we attempted to seek acidity-induced pro-apoptotic factors exerting a possible role as a sensitizer, and found that acidic condition elevates TNF-related apoptosis-inducing ligand (TRAIL) receptors, TRAIL-R1 (DR4) and TRAIL-R2 (DR5) in GC cells.
TRAIL receptors, members of the TNF receptor superfamily are membrane death receptors mediating TRAIL-induced apoptosis [12]. TRAIL is endogenous antitumor molecule and considered to be an attractive therapeutic agent since it is a potent and selective apoptotic inducer in transformed cells. TRAIL, secreted from natural killer cells initiates apoptosis by binding to and thus, engaging DR4 or DR5 expressed on the surface of tumor cells [12131415]. Following recruitment of the adaptor Fas-associated death domain (FADD) and initiator procaspase-8 to the death domains in the cytoplasmic tails of TRAIL receptors, the initiator is cleaved, resulting in onset of extrinsic or intrinsic caspase cascade, and finally triggering apoptosis [16].
In the present study, we observed that low pH contributes to TRAIL sensitivity and suggest a useful action of TRAIL to improve the effectiveness of GC treatment under acidic environment.
SNU-601 and AGS human GC cells were obtained from the Korean Cell Line Bank (Seoul, Korea) and American Type Culture Collection (Manassas, VA), respectively. Cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) fetal bovine serum and 1% PS at 37℃ in an atmosphere containing 5% CO2. Recombinant human tumor necrosis factor (TNF)-related apoptosis-inducing ligand (rhTRAIL) was a gift from T. H. Kim of the Department of Biochemistry and Molecular Biology, Chosun University, Korea [17]. Oxaliplatin was obtained from Boryung Pharmaceutical (Seoul, Korea), and cisplatin was purchased from Sigma-Aldrich (St. Louis, MO). Unless specified otherwise, drugs were purchased from Calbiochem (San Diego, CA).
Cells were incubated in growth medium adjusted to pH 7.4, 6.8, 6.5, or 6.2 for the indicated times (48 h if unspecified). To quantify growth, cells were collected at each time point and counted with a hemocytometer after being suspended and mixed with trypan blue. A fluorometric method was used to identify apoptotic cells. Briefly, treated cells were stained with 1 µg/ml Hoechst 33342 for 15 min, and both suspended and attached cells were collected and centrifuged. The pooled cell pellets were washed and fixed in 3.7% formaldehyde, and an aliquot of the suspension was centrifuged in a Cytospin cytocentrifuge (Thermo Fisher Scientific, Waltham, MA) for preparation of slides, which were then observed under a fluorescence microscope (DM5000, Leica, Wetzlar, Germany) using excitation and emission wavelengths of 340 and 425 nm, respectively. Condensed or fragmented nuclei were considered indicative of apoptosis. A total of 500 cells distributed across random microscope fields of view were counted, and the number of apoptotic and non-apoptotic cells was expressed as a percentage of the total. To detect necrotic cell death, LDH release was quantified using an LDH Cytotoxicity Assay Kit II (BioVision, Mountain View, CA) according to the manufacturer's protocol, as described previously [18].
EZ-cytox viability assay was performed as followed by manufacturer's protocol. Briefly, cells were plated in the wells of a 96-well plate at a density of 1−2×104 cells/well, cultured for 24 h, and then incubated in the growth medium adjusted to pH 7.4 or pH 6.5 for 48 h with combinatorial treatment of various drugs. The EZ-cytox solution (Daeillab, Korea) was added to the wells and incubated at 37℃ in a CO2 incubator for the last 2 h of incubation, and the plates were read using an enzyme-linked immunosorbent assay plate reader at 405 nm. The absorbance of the untreated cells was set as 100% and cell survival was expressed as a percentage of this value.
Treated cells were lysed in either a whole-cell lysis buffer (50 mM Hepes, 150 mM Nacl, 1% Triton X-100, 5 mM EGTA, protease inhibitor cocktail), or non-mitochondrial cytosolic buffer (for released cytochrome C, 75 mM NaCl, 8 mM Na2HPO4, 1 mM NaH2PO4, 250 mM sucrose, 1 mM EDTA, 350 µg/ml digitonin), and equal amounts of protein extracts were electrophoretically separated using 10–12% SDS-PAGE and transferred to a nitrocellulose membrane using a standard technique. Antibodies were used to probe for DR4, DR5 (ProSci, CA), Fas (Cell Signaling Technology, MA), and cytochrome C (Santa Cruz Biotechnology, CA). Anti-α-tubulin (BioGenex, CA) was used as a loading control. Signals were acquired using an Image Station 4000MM image analyzer (Kodak, NY).
For the RNAi experiment, siRNA of DR4, 5′-CUGGAAAGUUCAUCUACUU(dtdt)-3′ (sense) and 5′-AAGUAAUGAACUUUCCAG(dtdt)-3′ (anti-sense), DR5, 5′-CAGACUUGGUGCCCUUUG(dtdt)-3′ (sense) and 5′-UCAAAGGGCACCAAGUCUG(dtdt)-3′ (anti-sense), 5′-control siRNA, 5′-CCUACGCCACCAAUUUCGU(dtdt)-3′ (sense) and 5′-ACGAAAUUGGUGGCGUAGG (dtdt)-3′ (anti-sense) were purchased from Bioneer (Daejeon, Korea). Cells were individually transfected with siRNA oligonucleotides using an Amaxa Transfection System™ (Basel, Switzerland) and grown for 24 h prior to the treatment.
Real-time PCR was performed with the Light Cycler 2.0 (Roche) using the Fast Start DNA Master SYBR Green I Kit (Roche). For verification of the correct amplification product, PCR products were analyzed on a 2% agarose gel stained with ethidium bromide. The sequences of the primers were as follows: for β-actin, 5′- for Fas, 5′-TGGCACGGAACACACCCTGAGG-3′ and 5′-GAGGGTCCAGATGCCCAGCATG-3, for DR5, 5′-GCCTCATGGACAATGAGATAAAGGTGGCT-3′ and 5′-CCAAATCTCAAAGTACGCACAAACGG-3, for DR4, 5′-ATGGCGCCACCACCAGCTAGAG-3′ and 5′-CCCGCCTCGTGGTTCAATCCTC-3, for cyclin D1, 5′-CAT GTT CGT GGC CTC TAA G-3′ and 5′-GGA GAG GAA GTG TTC AAT GAA A-3′, for p21, 5′-CGATGGAACTTCGACTTTGTCAC-3′ and 5′-GGG CTT CCT CTT GGA GAA GAT CA-3′ for atg5, 5′-TGA GAT ATG GTT TGA ATA TGA AGG C-3′ and 5′-AGC TTC AAT TGC ATC CTT AGA-3′ for HSPA1B, 5′-CGACCTGAACAAGAGCAT-3′ and 5′-GATCAGGGCAGTCATCAC-3′, for COL13A1, 5′-TGGACCAAGTGGACCTC-3′ and 5′-TCACCCTTCATCCCAGC-3′, for COL20A1, 5′-ACCTATGTGTCCAGCGA-3′ and 5′-GGAGCTTTCTTGGTGGTC-3′, for slug, 5′-TTTGCCTCTCTTGCTTTGTTT-3′ and 5′-TAGCAGATGCAGCTAATGTTG A-3′, for snail, 5′-GGCTCCTTCGTCCTTCT-3′ and 5′-GGCTGAGGTATTCCTTGTT-3′ and for zeb1 5′-GCACTTGTCTTCTGTGTGA-3′ and 5′-ACTAGGAGACAAATTGCCAT-3′. Melting curve analysis was performed to confirm production of a single product. Negative controls without template were produced for each run. Data were analyzed using Light Cycler software version 4.0 (Roche, Switzerland).
The caspase-8 activity assay was carried out using a FADD-like IL-1β-converting enzyme (FLICE) colorimetric assay kit (BioVision), according to the manufacturer's protocol. Briefly, 200 µg protein lysates in a 50-µl volume was mixed with reaction buffer, mixed with IETD-pNA substrate, incubated for 90 min and the absorbance at 405 nm was measured. Fold increase in FLICE activity was determined by comparing the results of treated samples with the level of the untreated control.
All numerical data are presented as the mean±SE. All data represent the results of at least three independent experiments. Student's t-test was used to evaluate the differences in means between control and treatment group, and one-way ANOVA was applied to analyze differences caused by gene silencing or combined treatment. Significant difference was assumed at p<0.05.
To clarify the effect of acidic environments on the growth of GC cells, SNU-601 and AGS cells were counted after having been cultured for up to 72 h in medium adjusted to a normal (7.4) or acidic pH (6.8, 6.5, or 6.2). Although environmental acidity has been shown to induce the proliferation of several cancer cell types, in the present study, the growth rate of both GC cell lines decreased as culture medium acidity increased (Figs. 1A and B). In order to establish whether this reduction in cell number was due to cell death, the occurrence of apoptotic and non-apoptotic death was detected by monitoring apoptotic body formation (Fig. 1C) and LDH release (Fig. 1D), respectively. As shown in Figs. 1C and D, acidic culture conditions down to pH 6.2 induced only a negligible increase in apoptotic and non-apoptotic death, despite significant declines in the numbers of cells of both lines. Cell cycle analysis by flow cytometry revealed an increased proportion of cells in the G1 phase in lower-pH cultures, implying that cell cycle arrest may have contributed to the decreased cell counts noted under acidic conditions (Fig. 1E).
Acidity is known to modify the expression of various genes, and the phenotype of tumor cells in acidic environments may be dependent on patterns of gene expression. Thus, better understanding of acidity-mediated gene regulation may facilitate the identification of effective therapeutic strategies for gastric cancers exposed to acidic conditions. In this study, we selected several genes involved in regulation of the cell cycle, cell viability, resistance to apoptosis, and cancer progression, and explored changes in their expression levels in AGS cells under acidic culture conditions using real-time polymerase chain reaction (PCR) (Fig. 2).
Expression of the genes encoding cyclin D1, which plays a central role in progression of the cell cycle, and p21, a cell cycle inhibitor, was suppressed and elevated, respectively, under acidic culture conditions (pH 6.5) compared to pH 7.4, implying suppression of cell cycle progression in acidic environments (Figs. 2A and B). Culture at acidic pH values also resulted in upregulation of genes encoding an inducer of autophagy (ATG5), a molecular chaperone (HSPA1B), and the extracellular matrix (ECM) components collagen XIII and collagen XXA1 (Figs. 2C–F). Induction of autophagy or heat shock protein (HSP) expression is often related to the inhibition of apoptosis in response to various stressors [19202122], and upregulation of collagen expression has been linked to mechanisms of cell resistance to cytotoxic conditions [2324]. Hence, these results may support an anti-apoptotic role for environmental acidity through various mechanisms. As reported in previous studies, acidity also led to raised expression of several genes related to metastasis, including SNAIL and ZEB1, which may be responsible for the heightened tumor malignancy observed in acidic environments, although SLUG mRNA levels were not significantly increased (Figs. 2G–I). Interestingly, however, it was found that low-pH conditions also upregulated apoptosis-inducing genes, including FAS, DR4, and DR5 (Figs. 2J–L). Therefore, the acidic microenvironment appears to stimulate both cell survival and death mechanisms, as seen in response to other stresses.
Environmental acidity has been shown to affect the responses of cancer cells to anticancer drugs. To confirm that this also applies to GC cells, we compared the cytotoxicity of various drugs towards SNU-601 cells at normal and acidic pH (6.5) using an EZ-cytox assay. It has been suggested that low extracellular pH hinders cellular uptake of weakly basic drugs. In keeping with this, the cytotoxicity of doxorubicin and daunorubicin, classified as weakly basic drugs, was significantly lower in acidic culture medium than medium of a normal pH (Figs. 3A and B). Furthermore, under our experimental conditions, the cytotoxicity of various drugs was lower in acidic culture medium regardless of their pH. The cytotoxic effect of the alkylating agents; cisplatin and oxaliplatin, the topoisomerase inhibitor etoposide and the weakly acidic drug 5-fluorouracil on GC cells was decreased in medium of pH 6.5 (Figs. 3C–F). Consistent with these results, caspase-3 cleavage and cytochrome c release, indicative of apoptosis, were also reduced in acidic culture condition (Figs. 3G–L). Thus, extracellular acidity appears to render tumor cells resistant to multiple types of chemotherapeutic agents, not only weakly basic drugs.
Our analysis of gene expression patterns under acidic conditions revealed elevated expression of pro-apoptotic membrane death receptors in GC cells cultured at pH 6.5, although acidity is thought to be associated with anti-apoptotic and drug resistance mechanisms. To further examine the effect of acidity on the expression of death receptors, transcriptional levels of FAS, DR4, and DR5 were assessed at various acidic pH values. SNU-601 and AGS cells were cultured for 48 h in normal growth medium (pH 7.4) or acidic medium adjusted to pH 6.8, 6.5, or 6.2, and mRNA levels were analyzed by real-time PCR. As shown in Figs. 4A–F, relative expression of FAS, DR4, and DR5 was increased in low-pH conditions. In particular, DR5 exhibited higher basal mRNA levels and its expression was considerably raised at low pH values. The protein levels of these death receptors were also measured by immunoblotting. Moderate and very low basal levels of DR5 and DR4 were noted, respectively. Under acidic conditions, levels of the former were strongly raised, whereas only a slight increase in those of the latter was observed. FAS protein was rarely detected under normal or acidic conditions (Figs. 4G–H).
Since exposure of GC cells to acidity elevated expression of DR4 and DR5 proteins, we explored whether the susceptibility of such cells to TRAIL-induced apoptosis was heightened in acidic environments. To this end, we pretreated GC cells with normal growth medium or media of various acidic pH values for 48 h, before exposing them to low concentrations of rhTRAIL for 6 h and subsequently assessing apoptotic body formation. Treatment with rhTRAIL at pH 7.4 only slightly elevated the rate of apoptosis, but rhTRAIL-induced apoptosis was enhanced under acidic conditions (Fig. 5). This promotion of apoptosis seemed to be at least partially caused by upregulation of DR5, since silencing of this death receptor partly but significantly reduced the TRAIL-associated increase in apoptosis, as detected by apoptotic body formation (Fig. 6A) and caspase-8 and -3 activation (Figs. 6C and D). However, DR4 knockdown had no significant effects on acidity-promoted rhTRAIL-induced apoptosis, as assessed in the same manner (Figs. 6A–D).
It has been suggested that environmental acidity is a crucial factor driving the development of incurable drug-resistant tumor phenotypes. In this study, we attempted to elucidate the biology of tumor cells under acidic extracellular conditions and sought a strategy to overcome the detrimental effects of acidity on cancer treatment. Extracellular acidity per se suppressed cell growth by arresting cell cycle progression at the G1 phase, but did not significantly increase cell death. However, it also significantly affected the cytotoxicity towards GC cells of multiple chemotherapeutic drugs, as documented in other cancers [5].
In general, chemoresistance is associated with multifactorial processes, such as inhibition of drug uptake, deletion or mutation of receptors, alteration of metabolic processes targeted by drugs, and suppression of cell death mechanisms.
Although acidic microenvironment was reported to hinder the uptake of weakly basic drugs, the cytotoxicity of all of the various chemotherapeutic drugs used in this study was reduced, regardless of their ionic status in aqueous solution. Thus, it appears that the reduced effectiveness of such drugs under low-pH conditions is not only caused by decreased drug uptake, but also by other factors, such as overexpression of cytoprotective proteins, including HSPs, or induction of cell cycle arrest [2526]. Indeed, extracellular acidity altered the expression of multiple genes associated with cell cycle regulation, apoptosis, and cancer malignancy in the present study. Decreased cyclin D1 and increased p21 mRNA levels suggested suppression of cell cycle progression, and this was confirmed by flow cytometric analysis, which revealed increased numbers of cells in the G1 phase. In addition, extracellular acidity led to elevated expression of genes encoding the autophagy inducer ATG5 and molecular chaperone HSPA1B. In many cases, induction of autophagy plays a role in preventing apoptosis [192021], and overexpression of molecular chaperones such as HSP72 is related to cell survival and resistance to different environmental stresses [22]. Acidity also resulted in upregulated expression of genes encoding the ECM components collagen XIII and collagen XXA1. Raised collagen expression in response to environmental conditions is known to heighten resistance to cell death [2324]. Therefore, these results appear to support the idea that environmental acidity exerts anti-apoptotic effects through various mechanisms. Acidity also induced expression of several genes associated with metastasis, including SNAIL and ZEB1 [2728], consistent with acidic environments exerting an oncogenic influence.
During the course of this study, it was found that low pH also caused upregulation of apoptosis-inducing membrane death receptors, including DR4 and DR5 both in mRNA and protein levels in GC cells. Death receptors are members of the TNF receptor superfamily, and are activated upon interaction with their ligand TRAIL [12]. TRAIL is currently being evaluated as a good candidate for use as an effective and safe therapeutic agent. Preclinical studies in mice have shown that TRAIL induced apoptosis of tumor xenografts without apparent toxicity [14]. Moreover, it showed less toxicity than other members of apoptosis-inducing death ligands such as TNFα and Fas Ligand, which cause severe side-effects such as endotoxin-induced septic shock and ERK1/2-related oncogenic activities [2930]. However, recent studies reveal that many primary cancer cells are resistant to TRAIL-induced apoptosis due to various mechanisms, hindering its use as an effective therapeutic agent [31]. Indeed, clinical trials did not prove efficacy of TRAIL-based therapy, such as administration of recombinant TRAIL or agonistic monoclonal antibodies targeting to the death receptors [32]. Thus, significant effort is currently ongoing to seek a strategy to reverse this resistance. One of common mechanisms of TRAIL resistance is improper regulation of the death receptor signaling. Hence, a combination of TRAIL or TRAIL receptor agonists with a sensitizer making up for the defect has been evaluated as a strategy to overcome this limitation. So far, various natural and synthetic compounds have been screened for the activity as a potential TRAIL sensitizer, and multiple candidates including quercetin, genistein, curcumin, andrographolide and alkyllysophospholipid have been identified [333435]. One of their action mechanisms was shown to be upregulation of the death receptor signaling either by suppressing the inhibitory factors, such as c-FLIP or by elevating the death receptor expression [3637]. Given this, we took notice of the ability of acidity to upregulate the death receptor expression, and explored whether this phenomenon influences TRAIL sensitivity. As expected, addition of rhTRAIL to acidic culture medium markedly promoted apoptosis of GC cells. The TRAIL-sensitizing effect of acidity was significantly affected by silencing of DR5 but not DR4, indicating that the major role in this effect was played by DR5. It may be due to the big difference in the expression levels of these death receptors.
On the other hand, this acidity-induced DR upregulation appears to be a tissue-specific mechanism, since it has previously been shown that extracellular acidity does not elevate DR expression in DU145 cells [38]. Interestingly, however, the researchers that made this observation noted that extracellular acidity does augment TRAIL-induced apoptosis, but through stimulation of the mitochondrial apoptotic pathway. Therefore, their report agrees with our present study, in that in both investigations, TRAIL enhanced the apoptosis of tumor cells exposed to acidic extracellular conditions, although the details of the mechanisms involved may differ.
Based on the findings of the present study, we propose that TRAIL represents a promising local therapeutic option for gastric cancers exposed to acidic environments, though further advanced studies are required.
Figures and Tables
Fig. 1
GC cell growth was inhibited in acidic culture medium.
(A, B) AGS and SNU-601 cells were cultured in growth medium adjusted to the indicated pH for 24, 48, or 72 h, and counted at each time point. (C–E) Cells were incubated for 48 h in normal growth medium (pH 7.4) or acidic medium adjusted to pH 6.8, 6.5, or 6.2. Then cells are collected and stained with Hoechst 33342 to assess apoptotic cell death (C) or cells are fixed and stained with propidium iodide and analyzed by fluorescence-activated cell sorting analysis (E). Cell-free culture supernatants were subjected to an LDH release assay (D). *p<0.05 vs. pH 7.4.

Fig. 2
Extracellular acidity altered the expression of various genes.
AGS cells were incubated for 48 h in growth medium adjusted to pH 6.5, and mRNA expression of the genes encoding cyclin D1 (A), p21 (B), ATG5 (C), HSP70 (D), collagen XIII (E), collagen XXA1 (F), SLUG (G), SNAIL (H), ZEB1 (I), FAS (J), DR4 (K), and DR5 (L) was analyzed by real-time PCR. *p<0.05, **p<0.01 vs. pH 7.4.

Fig. 3
Extracellular acidic conditions decreased GC cell sensitivity to various chemotherapeutic drugs.
SNU-601 cells were cultured for 24 h in growth medium adjusted to pH 7.4 or 6.5, and subsequently exposed for 48 h to the indicated concentration of doxorubicin, daunorubicin, oxaliplatin, cisplatin, etoposide, or 5-fluorouracil in each pH-adjusted medium. The cells were then subjected to an EZ-cytox assay for measurement of cell viability (A–F), or immunoblotting of total protein extracts (for caspase-3 and α-tubulin) or cytosolic protein extracts (for released cytochrome c) (G–L).

Fig. 4
Expression of TRAIL-Rs in GC cells was upregulated in acidic culture conditions.
AGS and SNU-601 cells were exposed for 48 h to growth medium adjusted to pH 7.4, 6.8, 6.5, or 6.2. The treated cells were collected and mRNA expression of the genes FAS, DR4, and DR5 was analyzed by real-time PCR (A–F) or total protein extracts were prepared by cell lysis and analyzed by immunoblotting with antibodies against the corresponding proteins and α-tubulin as a loading control (G, H). *p<0.05, **p<0.01 vs. pH 7.4.
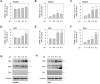
Fig. 5
Acidic culture conditions sensitized GC cells to rhTRAIL-induced apoptosis.
AGS (A, B) and SNU-601 (C, D) cells were exposed to normal growth medium (pH 7.4) or acidic medium (pH 6.5) for 48 h, before addition of 2 or 4 ng rhTRAIL and incubation for a further 6 h. The cells were then stained with Hoechst 33342, and images were captured under a fluorescence microscope (A, C) or apoptotic bodies were counted (B, D). The number of apoptotic cells is expressed as a percentage of the total number of cells counted. #p<0.05 vs. pH 7.4.

Fig. 6
TRAIL-R2/DR5 plays an essential role in acidity-promoted rhTRAIL-induced apoptosis.
(A, B) SNU-601 cells transfected with scrambled small interfering RNA (CTL RNAi) were exposed to normal growth medium (pH 7.4) and those transfected with CTL RNAi, DR5 RNAi, or DR4 RNAi were exposed to acidic culture medium (pH 6.5) for 42 h, before addition of 2 ng rhTRAIL and incubation for a further 6 h. The treated cells were then stained with Hoechst 33342, and apoptotic bodies were counted under a fluorescence microscope (A). The silencing effect of each small interfering RNA was confirmed by immunoblotting analysis of cells not administered rhTRAIL (B). (C, D) SNU-601 cells transfected with CTL RNAi, DR5 RNAi, or DR4 RNAi were exposed to growth medium at pH 6.5 for 42 h, before addition of 2 ng rhTRAIL and incubation for a further 4 h. The treated cells were subjected to a caspase-8 assay (C) and -3 activity assay (D). #p<0.05 vs. CTL RNAi- and rhTRAIL-treated cells.
ACKNOWLEDGEMENTS
This work was supported by a grant from the Clinical Medicine Research Institute of the Chosun University Hospital (2014). We thank Prof. Tae-Hyoung Kim for the kind gift of rhTRAIL and Ms. Jeong-Eun Choi for her excellent technical assistance.
Notes
References
1. Poole-Wilson PA. Measurement of myocardial intracellular pH in pathological states. J Mol Cell Cardiol. 1978; 10:511–526.

2. Reichert M, Steinbach JP, Supra P, Weller M. Modulation of growth and radiochemosensitivity of human malignant glioma cells by acidosis. Cancer. 2002; 95:1113–1119.

3. Thews O, Gassner B, Kelleher DK, Schwerdt G, Gekle M. Impact of hypoxic and acidic extracellular conditions on cytotoxicity of chemotherapeutic drugs. Adv Exp Med Biol. 2007; 599:155–161.

4. Wilting RH, Dannenberg JH. Epigenetic mechanisms in tumorigenesis, tumor cell heterogeneity and drug resistance. Drug Resist Updat. 2012; 15:21–38.

6. Fais S, De Milito A, You H, Qin W. Targeting vacuolar H+-ATPases as a new strategy against cancer. Cancer Res. 2007; 67:10627–10630.
7. Taylor S, Spugnini EP, Assaraf YG, Azzarito T, Rauch C, Fais S. Microenvironment acidity as a major determinant of tumor chemoresistance: Proton pump inhibitors (PPIs) as a novel therapeutic approach. Drug Resist Updat. 2015; 23:69–78.

8. Bertuccio P, Chatenoud L, Levi F, Praud D, Ferlay J, Negri E, Malvezzi M, La Vecchia C. Recent patterns in gastric cancer: a global overview. Int J Cancer. 2009; 125:666–673.

9. Peleteiro B, Severo M, La Vecchia C, Lunet N. Model-based patterns in stomach cancer mortality worldwide. Eur J Cancer Prev. 2014; 23:524–531.

10. Deng K, Lin S, Zhou L, Li Y, Chen M, Wang Y, Li Y. High levels of aromatic amino acids in gastric juice during the early stages of gastric cancer progression. PLoS One. 2012; 7:e49434.

11. Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg JM, Sloane BF, Johnson J, Gatenby RA, Gillies RJ. Acidity generated by the tumor microenvironment drives local invasion. Cancer Res. 2013; 73:1524–1535.

12. Wang S. The promise of cancer therapeutics targeting the TNF-related apoptosis-inducing ligand and TRAIL receptor pathway. Oncogene. 2008; 27:6207–6215.

13. Gura T. How TRAIL kills cancer cells, but not normal cells. Science. 1997; 277:768.
14. Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999; 5:157–163.

15. Kashii Y, Giorda R, Herberman RB, Whiteside TL, Vujanovic NL. Constitutive expression and role of the TNF family ligands in apoptotic killing of tumor cells by human NK cells. J Immunol. 1999; 163:5358–5366.
16. Baker SJ, Reddy EP. Modulation of life and death by the TNF receptor superfamily. Oncogene. 1998; 17:3261–3270.

17. Shin JN, Park SY, Cha JH, Park JY, Lee BR, Jung SA, Lee ST, Yun CW, Seol DW, Kim TH. Generation of a novel proform of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) protein that can be reactivated by matrix metalloproteinases. Exp Cell Res. 2006; 312:3892–3898.

18. Lim SC, Choi JE, Kang HS, Han SI. Ursodeoxycholic acid switches oxaliplatin-induced necrosis to apoptosis by inhibiting reactive oxygen species production and activating p53-caspase 8 pathway in HepG2 hepatocellular carcinoma. Int J Cancer. 2010; 126:1582–1595.

19. Luo S, Rubinsztein DC. Atg5 and Bcl-2 provide novel insights into the interplay between apoptosis and autophagy. Cell Death Differ. 2007; 14:1247–1250.

20. Apel A, Herr I, Schwarz H, Rodemann HP, Mayer A. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 2008; 68:1485–1494.

21. Han W, Sun J, Feng L, Wang K, Li D, Pan Q, Chen Y, Jin W, Wang X, Pan H, Jin H. Autophagy inhibition enhances daunorubic-ininduced apoptosis in K562 cells. PLoS One. 2011; 6:e28491.

22. Buzzard KA, Giaccia AJ, Killender M, Anderson RL. Heat shock protein 72 modulates pathways of stress-induced apoptosis. J Biol Chem. 1998; 273:17147–17153.

23. Su C, Su B, Tang L, Zhao Y, Zhou C. Effects of collagen iv on cisplatin-induced apoptosis of non-small cell lung cancer cells. Cancer Invest. 2007; 25:542–549.

24. Januchowski R, Świerczewska M, Sterzyńska K, Wojtowicz K, Nowicki M, Zabel M. Increased expression of several collagen genes is associated with drug resistance in ovarian cancer cell lines. J Cancer. 2016; 7:1295–1310.

25. Vukovic V, Tannock IF. Influence of low pH on cytotoxicity of paclitaxel, mitoxantrone and topotecan. Br J Cancer. 1997; 75:1167–1172.

26. Wachsberger PR, Landry J, Storck C, Davis K, O'Hara MD, Owen CS, Leeper DB, Coss RA. Mammalian cells adapted to growth at pH 6.7 have elevated HSP27 levels and are resistant to cisplatin. Int J Hyperthermia. 1997; 13:251–255. discussion 257-9.

27. Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009; 15:195–206.

28. Tan X, Banerjee P, Liu X, Yu J, Gibbons DL, Wu P, Scott KL, Diao L, Zheng X, Wang J, Jalali A, Suraokar M, Fujimoto J, Behrens C, Liu X, Liu CG, Creighton CJ, Wistuba II, Kurie JM. The epithelial-to-mesenchymal transition activator ZEB1 initiates a prometastatic competing endogenous RNA network. J Clin Invest. 2018; 128:1267–1282.

29. Hardy J, Jones A, Gore ME, Viner C, Selby P, Wiltshaw E. Treatment of advanced ovarian cancer with intraperitoneal tumour necrosis factor. Eur J Cancer. 1990; 26:771.

30. Barnhart BC, Legembre P, Pietras E, Bubici C, Franzoso G, Peter ME. CD95 ligand induces motility and invasiveness of apoptosis-resistant tumor cells. EMBO J. 2004; 23:3175–3185.

31. Srivastava RK. TRAIL/Apo-2L: mechanisms and clinical applications in cancer. Neoplasia. 2001; 3:535–546.

32. Lemke J, von Karstedt S, Zinngrebe J, Walczak H. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014; 21:1350–1364.

33. Lim SC, Parajuli KR, Han SI. The alkyllysophospholipid edelfosine enhances TRAIL-mediated apoptosis in gastric cancer cells through death receptor 5 and the mitochondrial pathway. Tumour Biol. 2016; 37:6205–6216.

34. Jin CY, Park C, Cheong J, Choi BT, Lee TH, Lee JD, Lee WH, Kim GY, Ryu CH, Choi YH. Genistein sensitizes TRAIL-resistant human gastric adenocarcinoma AGS cells through activation of caspase-3. Cancer Lett. 2007; 257:56–64.

35. Jung EM, Park JW, Choi KS, Park JW, Lee HI, Lee KS, Kwon TK. Curcumin sensitizes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis through CHOP-independent DR5 upregulation. Carcinogenesis. 2006; 27:2008–2017.

36. Yoon MJ, Kang YJ, Kim IY, Kim EH, Lee JA, Lim JH, Kwon TK, Choi KS. Monensin, a polyether ionophore antibiotic, overcomes TRAIL resistance in glioma cells via endoplasmic reticulum stress, DR5 upregulation and c-FLIP downregulation. Carcinogenesis. 2013; 34:1918–1928.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download