Abstract
Background and Purpose
Methods
Results
Figures and Tables
Fig. 1
Quantile-quantile plots of the association results for the initial genome scan. The tail of the distribution of gene-based probability values deviated more significantly than those of SNPs inside or outside of the gene, which suggests that the power was higher for a gene-based association analysis than a single SNP-based GWAS in detecting associations. GWAS: genome-wide association study, SNP: single-nucleotide polymorphism.

Fig. 2
Manhattan plot of gene probability values on chromosomes. Most of the genes significantly associated with MS are mapped to the HLA region (chromosome 6). HLA: human leukocyte antigen, MS: multiple sclerosis.

Fig. 3
Protein-protein interactions between MS-associated genes. Only the connected genes are shown. The novel genes are labeled in red. Different line colors represent the types of evidence for the association. HLA class I cluster: major histocompatibility complex, class I, A (HLA-A), Major histocompatibility complex, class I, C (HLA-C), and major histocompatibility complex, class I, F (HLA-F); HLA class II cluster: major histocompatibility complex, class II, DQ alpha 1 (HLA-DQA1), major histocompatibility complex, class II, DQ alpha 2 (HLA-DQA2), major histocompatibility complex, class II, DQ beta 2 (HLA-DQB2), major histocompatibility complex, class II, DR alha (HLA-DRA), major histocompatibility complex, class II, DO beta (HLA-DOB), major histocompatibility complex, class II, DM alpha (HLA-DMA), and major histocompatibility complex, class II, DM beta (HLA-DMB). HLA: human leukocyte antigen, MS: Multiple sclerosis.
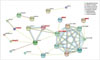
Table 1
MS-associated genes with significantly differential expressions in MS-related cells/tissue

The ratio listed in the line of "Sample size" is the number of multiple sclerosis (MS) cases compared with that of controls. The genes listed here that are not defined in the main text are major histocompatibility complex, class II, DO beta (HLA-DOB), major histocompatibility complex, class II, DR alha (HLA-DRA), POU class 5 homeobox 1 (POU5F1), uncharacterized LOC100294145 (LOC100294145), tenascin XB (TNXB), psoriasis susceptibility 1 candidate 1 (PSORS1C1), major histocompatibility complex, class II, DM beta (HLA-DMB), transporter 2, ATP-binding cassette, sub-family B (TAP2), allograft inflammatory factor 1 (AIF1), major histocompatibility complex, class I, F (HLA-F), major histocompatibility complex, class II, DQ alpha 1 (HLA-DQA1), respectively.
*Only the most significant probe is listed, even if more than one probe was detected for a gene.
GSE No.: Gene Expression Omnibus number, www.ncbi.nlm.nih.gov/geo/, PMID: PubMed unique identifier.
Table 2
Fifteen novel MS-associated genes, supported by replication studies and/or expression studies

The genes listed here that are not defined in the main text are tripartite motif containing 26 B (TRIM26P), HLA complex P5 pseudogene 14 (HCP5P14), HLA complex group 26 pseudogene (3.8-1.4), mucin 21, cell surface associated (MUC21), HLA-F antisense RNA 1 (HLA-F-AS1), respectively.
*Number of SNPs included in the gene with p<0.05, †Multiple sclerosis (MS)-related cells/tissue sample used for the differential expression analysis; S1, PBMC; S2, CD34+ HPC; S3, CD8+ T lymphocytes; S4, spinal cord.
NA: not available, NS: not significant.
Acknowledgements
References
Supplementary Materials
Supplementary Table 1
Results of gene-based and differential expression analyses of 58 MS-associated genes

PheGenI, Phenotype-Genotype Integrator.
*The number of SNP included in a gene with p-value<0.05, †Ms-related cells sample used to differential expression ananlysis. S1: PBMC, S2: CD34+ HPC, S3: CD8+ T lymphocytes, S4, spinal cord.
The genes listed here that are not defined in the main text are chromosome 6 open reading frame 10 (C6orf10), notch 4 (NOTCH4), small nucleolar RNA, H/ACA box 38 (SNORA38), proline-rich coiled-coil 2A (BAT2), BCL2-associated athanogene 6 (BAT3), peptidylprolyl isomerase A pseudogene 9 (PPIAP9), major histocompatibility complex, class II, DQ beta 2 (HLA-DQB2), major histocompatibility complex, class II, DQ alpha 2 (HLA-DQA2), ubiquitin specific peptidase 8 pseudogene 1 (LOC100287272), mitochondrial coiled-coil domain 1 (MCCD1), fibroblast growth factor receptor 3 pseudogene 1 (LOC100462812), HLA complex group 27 (HCG27), major histocompatibility complex, class II, DR beta 9 (HLA-DRB9), major histocompatibility complex, class I, S (HLA-S), butyrophilin-like 2 (BTNL2), DEAH (Asp-Glu-Ala-His) box polypeptide 16 (DHX16) transcription factor 19 (TCF19), HLA complex group 26 pseudogene (3.8-1.5), MHC class I polypeptide-related sequence A (LOC100507436), major histocompatibility complex, class I, x (HLA-X), HLA complex group 4 pseudogene 4 (HCG4P4), ribosomal protein L3 pseudogene 2 (RPL3P2), dihydrofolate reductase pseudogene 2 (DHFRP2), MHC class I polypeptide-related sequence E (MICE), ripartite motif containing 31 (TRIM31), MAX dimerization protein 3 (MXD3), similar to hCG1987428 (LOC100287247), meiotic nuclear divisions 1 homolog (LOC100127934), ribosomal protein L15 pseudogene 4 (RPL15P4), HLA complex group 2 pseudogene 8 (HCG2P8) respectively.
Supplementary Table 2
Enrichment of GO terms and KEGG pathways of the 58 MS-associated genes

Count, the number of genes enriched in particular GO terms or KEGG pathways; Genes, potential genes enriched in particular GO terms or KEGG pathways.
*p value with Bonferroni correction for multiple tests. Only significant results with p<0.05 are listed.
GO: Gene Ontology, KEGG: Kyoto Encyclopedia of Genes and Genomes, MHC: major histocompatibility complex, MS: mltiple sclerosis.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download