Abstract
Background
Methods
Results
Acknowledgments
Notes
Authors' Contributions: Conceptualization: Lee KH. Methodology: Yun YJ, Lee JS, Yoo JC, Cho E, Park D. Formal analysis: Lee KH, Yun YJ, Lee JS, Yoo JC, Park D. Data curation: Yun YJ, Lee JS, Yoo JC, Lee KH, Cho E, Park D. Validation: Lee KH, Yun YJ, Lee JS, Yoo JC, Kook YH. Investigation: Lee KH. Writing-original draft preparation: Yun YJ, Lee KH. Writing-review and editing: Yun YJ, Lee JS, Yoo JC, Lee KH. Approval of final manuscript: all authors.
References
Supplementary Material
Table 1
Drug resistance profiles of 93 Mycobacterium tuberculosis isolates
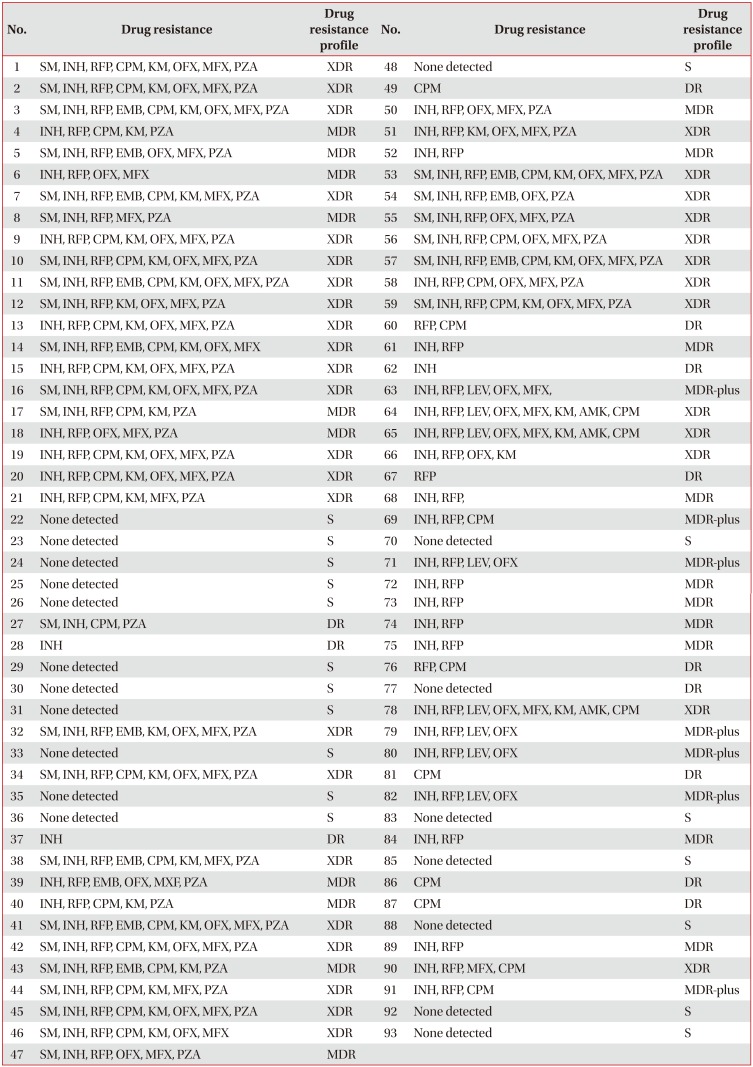
SM: streptomycin; INH: isoniazid; RFP: rifampicin; CPM: capreomycin; KM: kanamycin; OFX: ofloxacin; MFX: moxifloxacin; PZA: pyrazinamide; EMB: ethambutol; XDR: extensively drug-resistant; MDR: multidrug-resistant; S: susceptibility to all of the drugs; DR: drug resistance other than MDR (including MDR-plus and XDR); MDR-plus: INH+RFP+fluoroquinolone or INH+RFP+injectable drugs.
Table 2
Isolates with rpoC mutations (n=24)
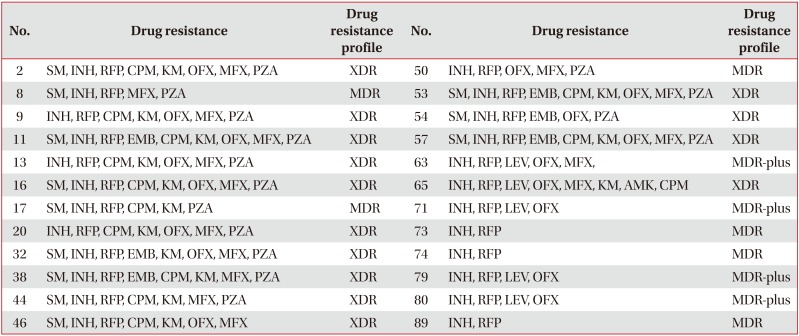
Isolates with mutations in rpoC are resistant to both INH and RFP.
SM: streptomycin; INH: isoniazid; RFP: rifampicin; CPM: capreomycin; KM: kanamycin; OFX: ofloxacin; MFX: moxifloxacin; PZA: pyrazinamide; EMB: ethambutol; XDR: extensively drug-resistant; MDR-plus: INH+RFP+fluoroquinolone or INH+RFP+injectable drugs.
Table 3
Mutations detected in the rpoC gene of 93 Mycobacterium tuberculosis isolates
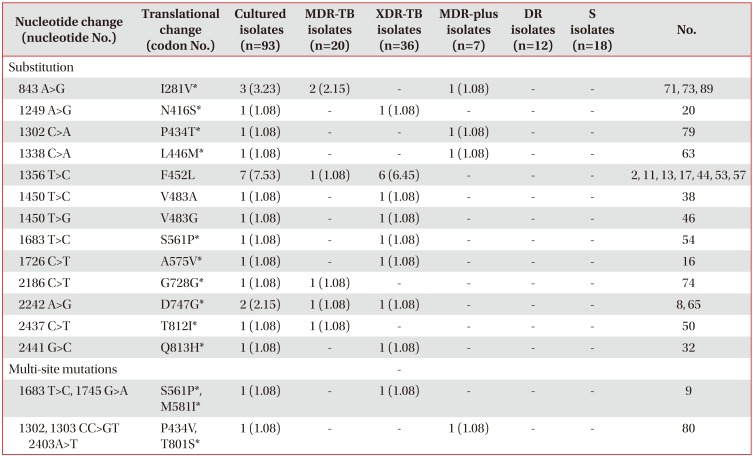
Values are presented as number (%).
*New mutation not reported in previous studies. Isolates with mutations in rpoC are resistant to both INH and RFP.
MDR-TB: multidrug-resistant tuberculosis; XDR-TB: extensively drug-resistant tuberculosis; MDR-plus: INH+RFP+fluoroquinolone or INH+RFP+injectable drugs; DR: drug resistance other than MDR (including MDR-plus and XDR); S: susceptibility to all of the drugs; INH: isoniazid; RFP: rifampicin.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download