Abstract
Background
Although testing to detect weak D antigens using the antihuman globulin reagent is not required for D− patients in many countries, it is routinely performed in Korea. However, weak D testing can be omitted in D− patients with a C−E− phenotype as this indicates complete deletion of the RHD gene, except in rare cases. We designed a new algorithm for weak D testing, which consisted of RhCE phenotyping followed by weak D testing in C+ or E+ samples, and compared it with the current algorithm with respect to time and cost-effectiveness.
Methods
In this retrospective study, 74,889 test results from January to July 2017 in a tertiary hospital in Korea were analyzed. Agreement between the current and proposed algorithms was evaluated, and total number of tests, time required for testing, and test costs were compared. With both algorithms, RHD genotyping was conducted for samples that were C+ or E+ and negative for weak D testing.
Results
The algorithms showed perfect agreement (agreement=100%; κ=1.00). By applying the proposed algorithm, 29.56% (115/389 tests/yr) of tests could be omitted, time required for testing could be reduced by 36% (8,672/24,084 min/yr), and the test cost could be reduced by 16.53% (536.11/3,241.08 USD/yr).
Weak D is defined as decreased reactivity (negative to 2+) of red blood cells (RBCs) with anti-D reagent, but moderate or strong agglutination in weak D testing [123]. Weak D testing is a method that detects weak expression of D, and its goal in RBC donors is identification of units with weak D or partial D types to prevent anti-D immunization of recipients. Weak D testing is time consuming and costly, as it involves the use of antiglobulin reagents and constant temperature control at 37℃. In Western countries, performing this test for all serologically weak D or D− individuals can substantially increase the workload of laboratory workers. This was the reason behind the American Association of Blood Bank (AABB) recommendation to exclude weak D testing for blood recipients and to reallocate resources to prevent alloimmunization of D− or D-variant women [45].
In contrast, in some countries such as Korea, Japan, and France and in 19.8% of the United States (US) medical institutions, weak D testing is routinely conducted on all samples from both blood donors and recipients when a D− result is suspected [4678]. Because the frequency of Korean D− blood donors is only 0.15% [9], we speculate that routine application of weak D testing for all serological weak D phenotype recipients does not pose an excessive additional burden on the clinical blood bank, while avoiding confusion about D type, in case these patients become blood donors. The most recent version of the Excellent Laboratory Certification in Korea (2018) maintains the policy that weak D testing is necessary for all D− recipients of transfusion [10]. RHD genotyping for the detection of Asia-type DEL (c.1227G>A) is also encouraged for D− recipients especially in Asian countries, as recipients with this mutation can safely receive a transfusion from D+ donors when there is a shortage of D− blood [1112].
Recently, Seo et al [12] reported an effective diagnostic strategy for accurate detection of D variants, including Asia-type DEL, in apparently D− blood donors in Korea. Based on the hypothesis that all D− samples with a C−E− phenotype have complete deletion of the RHD gene, they concluded that RHD genotyping is not necessary in D− cases with a C−E− phenotype. In line with that study, we hypothesized that weak D testing is not necessary in D− samples with a C−E− phenotype, as they are speculated to have a complete deletion of the RHD gene.
In order to propose a time and cost-effective D testing strategy for Korea, we designed a new algorithm for weak D testing and compared the current and new proposed algorithms in terms of time and cost-effectiveness in a tertiary hospital.
This retrospective study was performed on 74,889 test results from both in-patients and outpatients between January and July 2017 at Samsung Medical Center, Seoul, Korea. This study was approved by the Institutional Review Board of Samsung Medical Center (SMC-2017-06-061). D typing grade 2 or weaker was included in the study. Patients' test results were collected using a laboratory information system. Patients' disease status did not affect enrollment. As the study used the retrospectively collected data in anonymity, written informed consent was waived.
Galileo NEO (Immucor, Rödermark, Germany) and QWALYS-3 (Diagast, Loos, France) were used for D typing using Novaclone (Immucor, Rödermark, Germany) and ABD-Lys (Diagast, Loos, France), respectively. D typing was duplicated using the manual tile method with the anti-D Bioclone (MAD2 clone; Ortho Clinical Diagnostics, Raritan, NJ, USA). Samples showing strong positive reactivity (grade 3+ to 4+) for both the automated and the manual tile tests were considered D+ and were not further tested [13]. Samples showing weak reactivity (agglutination strength of grade 2+ or weaker) with one or more D typing reagents used in an automated instrument and manual D typing were considered D-variants, and negative samples were subjected to weak D testing and/or RhCE phenotyping. RhCE phenotyping was conducted using the immediate-spin tube test with monoclonal anti-C, −c, −E, and −e (Ortho Clinical Diagnostics, High Wycombe, UK) in saline-filled test tubes. The monoclonal/polyclonal blend of anti-D and human IgG/IgM monoclonal anti-D (Millipore, Livingston, UK) was used for weak D testing. Samples with a C−E− phenotype were considered D−. RHD genotyping was done by screening four loci of the RHD gene (promoter, exon 4, intron 7, and exon 10) by PCR amplification. Direct sequencing was done on all 10 exons. The methods used have been described in detail previously [12]. RHD genotyping discriminated between D-variants, including Asia-type DEL (c.1227G>A) and D−.
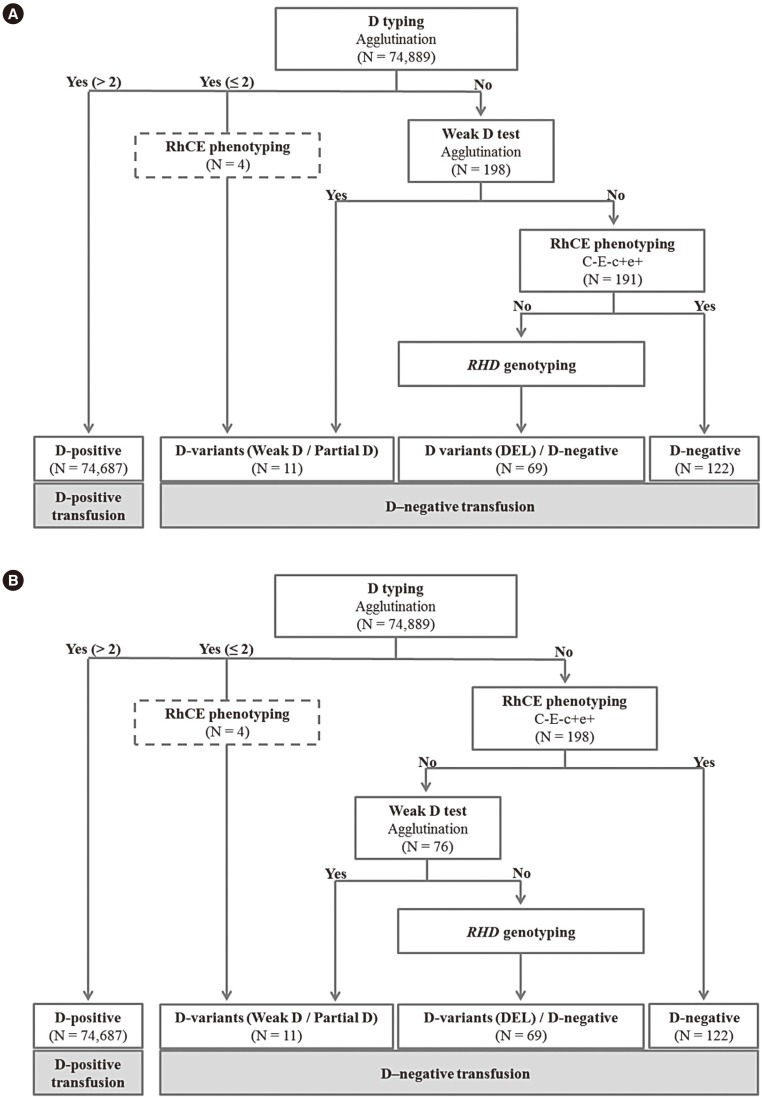
The current and proposed algorithms for D typing are shown in Fig. 1. Using the current algorithm, weak D testing was first conducted on patients who show no response to routine D testing. Then, RhCE phenotyping was performed for samples that were negative on weak D testing (Fig. 1A). In the proposed algorithm, first, RhCE phenotyping was carried out for patients with a negative response in the routine D testing, which was followed by weak D testing on non-C−E− samples (Fig. 1B). Weak D-negative patients were subjected to RHD genotyping using both algorithms.
The average monthly incidence of patients with grade 2 or weaker D typing was calculated, and annual incidence was estimated. The total time and cost of each algorithm were calculated for a month or a year by multiplying the number of required tests by the time and cost of each test. Cost was calculated according to the reimbursement datasheet provided by the Health Insurance Review and Assessment Service of Korea (published in March 2017), the costs of the weak D testing and RhCE phenotyping were set at 3.04 and 7.03 USD, respectively [14]. Time was calculated by receipt-to-verification turnaround time (TAT) of each test. To evaluate the TAT, each of the two tests was performed 20 times and mean and standard deviation were calculated.
All analyses were performed using Analyse-it 4.92.4 (Analyse-it Software Ltd., Leeds, UK). We evaluated the agreement of the D status classification in both algorithms using Cohen's kappa statistic. For interpreting κ values, we used the criteria set by Landis and Koch [15]: <0=poor agreement; 0.01 to 0.20=slight agreement; 0.21 to 0.40=fair agreement; 0.41 to 0.60=moderate agreement; 0.61 to 0.80=substantial agreement; and 0.81 to 0.99=nearly perfect agreement.
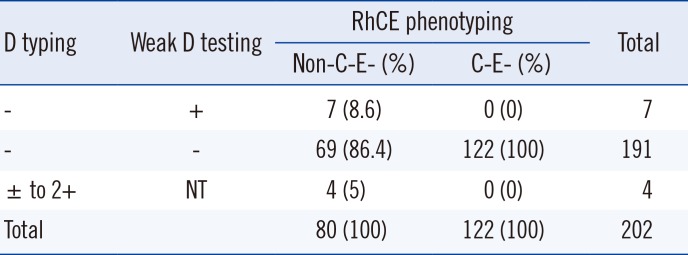
A total 198 samples were D−, and four samples showed trace to 2+ reactivity in D typing. RhCE phenotyping results of all 202 samples are summarized in Table 1. Seven D− samples were positive in weak D testing, and 122 D− samples with the C−E− phenotype were all negative on weak D testing. None of the C−E− phenotype samples were positive for weak D testing, with perfect agreement (202/202, agreement=100%; κ=1.00) between the two algorithms (Table 1). The remaining 74,687 samples showed higher reactivity than 2+ in D typing and were classified as D+.
The average monthly and estimated annual patient incidence was (mean±SD) 29±7 and 345±43.5 persons, respectively. With the proposed algorithm, 122 weak D tests could be omitted while seven RhCE phenotyping tests would be added during this study period. The analysis time for weak D testing and RhCE phenotyping was (mean±SD) 44.35±1.18 and 29.68±8.19 minutes, respectively. The time required for testing was reduced by 36% (5,203/14,450 minutes), and the test cost was calculated to save 16.53% (321.67/1,944.65 USD).
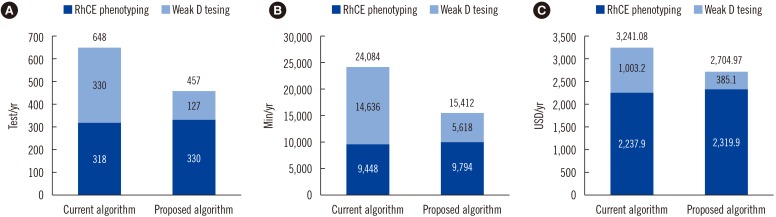
Based on the study results, the number of tests, time required for testing, and test cost of each algorithm performed for a period of one month are as follows (new/old algorithm): 40/57 tests, 1,360/2,125 minutes, and 238.67/285.98 USD, respectively. The values calculated for one year for the two test algorithms are as follows (new/old algorithm): 457/648 tests, 15,412/24,084 minutes, and 2,704.97/3,241.08 USD, respectively (Fig. 2A, 2B, and 2C).
Recently, the AABB-College of American Pathologists (CAP) Work Group recommended that RHD genotyping be carried out for transfusion recipients when a serological weak D phenotype is detected by routine D typing [16]. Those patients whose serological weak D phenotype is associated with molecularly defined weak D type 1, 2, or 3 can be transfused safely with D+ RBCs [217]. This can be especially useful for patients requiring chronic transfusions. This recommendation can be extrapolated to Asia-type DEL recipients, who can safely receive transfusion from D+ donors. RHD genotyping for clinical purposes is currently conducted only for the detection of Asia-type DEL in D− samples showing a C+ or E+ phenotype in our institution. RHD genotyping is currently not conducted on weak D samples, as the incidence of weak D type 1, 2, or 3 is very low in the Korean population (unpublished data). In this study, only 28.9% (22/76) of D− patients with a C+ or E+ phenotype, including Cce, cDe, CcEe, cEe, and Ce, were subjected to RHD genotyping. Nine serologically D− patients were identified as Asia-type DEL.
The frequency of the DEL phenotype is much higher among Cde and cdE haplotypes than among cde haplotypes [9]. DEL type with C−E− phenotype has been rarely reported. Flegel et al [18] reported an RHD (634G>C) blood donor among 43,053 C−E− phenotype donors in Germany. In Asia, two samples with DEL type with C−E− phenotype has been reported in the Japanese [19] and Chinese [20]. Therefore, the introduction of our new algorithm to evaluate blood donors requires sufficient consideration. However, the purpose of this study was to maximize the efficiency of D typing for patients who are prepared for transfusion therapy in hospitals. The major significance of RHD genotyping in patients is the identification of Asia-type DEL patients, who can receive D+ blood.
Our group previously reported an effective diagnostic strategy omitting RHD genotyping in D− cases with a C−E− phenotype, as the D− phenotype in these cases is caused by complete deletion of the RHD gene, except in extremely rare cases [12]. In the current study, we applied the same hypothesis to weak D testing and showed that this test is not necessary for all D− samples with a C−E− phenotype. Our retrospective study comparing the current and newly proposed algorithms showed that both algorithms agreed perfectly.
Omitting weak D testing, according to RhCE phenotyping, was found to be efficient without any difference in test results. Thus, we conclude that weak D testing can be omitted in C−E−c+e+ phenotype cases. The improved algorithm is expected to have a significant impact on the operation of blood banks by reducing the time required for testing by 36% and annual test costs by 16.53%.
As this study was limited by the small number of D− patients recruited and was a single center study, further larger-scale studies are warranted to verify our findings. Despite this limitation, application of our algorithm in blood centers will improve time and cost-effectiveness.
Acknowledgment
This study was supported by research funds provided by the Laboratory Medicine Foundation, Seoul, Korea.
References
1. Sandler SG, Flegel WA, Westhoff CM, Denomme GA, Delaney M, Keller MA, et al. It's time to phase in RHD genotyping for patients with a serologic weak D phenotype. College of American Pathologists Transfusion Medicine Resource Committee Work Group. Transfusion. 2015; 55:680–689. PMID: 25438646.
2. Daniels G. Variants of RhD--current testing and clinical consequences. Br J Haematol. 2013; 161:461–470. PMID: 23432139.
3. Jenkins CM, Johnson ST, Bellissimo DB, Gottschall JL. Incidence of weak D in blood donors typed as D positive by the Olympus PK 7200. Immunohematology. 2005; 21:152–154. PMID: 16472016.
4. Sandler SG, Roseff SD, Domen RE, Shaz B, Gottschall JL. Policies and procedures related to testing for weak D phenotypes and administration of Rh immune globulin: results and recommendations related to supplemental questions in the Comprehensive Transfusion Medicine survey of the College of American Pathologists. Arch Pathol Lab Med. 2014; 138:620–625. PMID: 24786120.
5. AABB. Standards for Blood Banks and Transfusion services. 18th ed. American Association of Blood Banks;2016. p. 327–328.
6. Han KS, Park KU, Song EY. Transfusion medicine. 4th ed. Seoul: Korea Medical Book Publishing Company;2014. p. 367–370.
7. Japan Society of Transfusion Medicine and Cell Therapy, Transfusion examination technology training course committee Ver.1.3.1. Inspection manual for blood transfusion. Tokyo: Japan Society of Transfusion Medicine and Cell Therapy, Transfusion examination technology training course committee;2017. p. 3–5.
8. Hors collection. Transfusion medicine: the French model. Montrouge: John Libbey Eurotext;2013. p. 61–82.
9. Kim JY, Kim SY, Kim CA, Yon GS, Park SS. Molecular characterization of D- Korean persons: development of a diagnostic strategy. Transfusion. 2005; 45:345–352. PMID: 15752151.
10. Laboratory Medicine Foundation. 2018 Checklist for Excellent Laboratory Certification_Transfusion Medicine. Updated on Dec 2017. http://lmf.or.kr/sub/catalog.
11. Shao CP. Transfusion of RhD-positive blood in “Asia type” DEL recipients. N Engl J Med. 2010; 362:472–473. PMID: 20130261.
12. Seo MH, Won EJ, Hong YJ, Chun S, Kwon JR, Choi YS, et al. An effective diagnostic strategy for accurate detection of RhD variants including Asian DEL type in apparently RhD-negative blood donors in Korea. Vox Sang. 2016; 111:425–430. PMID: 27864976.
13. Lim YA, Cho HS, Choi YS, Jang CH, Lee MN, Kwon JR, et al. Report on external proficiency testing for the ABO and D blood group typing tests in blood centers (2015). Korean J Blood Transfus. 2016; 27:68–78.
14. Korean Medical Association. Health insurance review and assessment service of Korea. Seoul: Kyungsung Media;2017. p. 110.
15. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977; 33:159–174. PMID: 843571.
16. Sandler SG, Chen LN, Flegel WA. Serological weak D phenotypes: a review and guidance for interpreting the RhD blood type using the RHD genotype. Br J Haematol. 2017; 179:10–19. PMID: 28508413.
17. Pham BN, Roussel M, Peyrard T, Beolet M, Jan-Lasserre V, Gien D, et al. Anti-D investigations in individuals expressing weak D Type 1 or weak D Type 2: allo- or autoantibodies? Transfusion. 2011; 51:2679–2685. PMID: 21658048.
18. Flegel WA, von Zabern I, Wagner FF. Six years' experience performing RHD genotyping to confirm D- red blood cell units in Germany for preventing anti-D immunizations. Transfusion. 2009; 49:465–471. PMID: 19243542.
19. Ogasawara K, Suzuki Y, Sasaki K, Osabe T, Isa K, Tsuneyama H, et al. Molecular basis for D- Japanese: identification of novel DEL and D- alleles. Vox Sang. 2015; 109:359–365. PMID: 25953588.
20. Wang M, Wang BL, Xu W, Fan DD, Peng ML, Pan J, et al. Anti-D alloimmunisation in pregnant women with DEL phenotype in China. Transfus Med. 2015; 25:163–169. PMID: 26033335.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download